Day 1: Data wrangling
Intro to the data set and class set up (15 min)
- We will be working with data from the Portal Project Teaching Database.
- All data is available on figshare.
- It is a simplified version of data from study Ernest etal. 2016. Long-term monitoring and experimental manipulation of a Chihuahuan Desert ecosystem near Portal, Arizona, USA.
- See Portal, Arizona in the map.
- The original database is published at Ecological Archives
- Long-term experimental study, showing a time-series for a small mammal community in southern Arizona. This is part of a project studying the effects of rodents and ants on the plant community that has been running for almost 40 years. The rodents are sampled on a series of 24 plots, with different experimental manipulations controlling which rodents are allowed to access which plots.
- 24 experimental and control plots
- Experimental manipulations over the years include removal of all or some rodent species, all or some ants, seed additions, and various alterations of the annual plant community.
- The dataset is composed of three tables, which includes information on the site, date, species identification, weight and sampling plot (within the site) for some small mammals in Arizona.
-
Each table is stored as a CSV file.
- CSV stands for “comma separated values”
- CSV is common way of storing data that can be used across programming and data management software
- If we look at one of these files we can see that
- It is plain text, so any program can read it
- The first row is the header row, with different column headers separated by commas
- All of the other rows are the data, again with different columns separated by commas
- And so each of the values is separated by commas, hence “comma separated values”
Setup your RStudio project
- Remember! Working in projects is considered a best practice in data science.
- Open your RStudio project. I called mine
data-science. - Click on the names of the following files to download them:
- surveys.csv - main table, one row for each rodent captured, date on date, location, species ID, sex, and weight in grams and hindfoot length in millimeters.
- species.csv - latin species names for each species ID + general taxon
- plots.csv - information on the experimental manipulations at the site
- Save the three CSV files into your “data-raw” folder.
- Don’t know where the CSV files are? Right click ->
Save link as.
- Don’t know where the CSV files are? Right click ->
- Create a new Rmd file and save it in the “documents” folder with the name “small-mammals.Rmd”.
- Use markdown syntax to create a description of this dataset.
Loading and viewing the dataset
- Load the three CSV files into
Rusing the functionread.csv()from the packageutils. - Remember to use relative paths.
surveys <- read.csv("surveys.csv")
species <- read.csv("species.csv")
plots <- read.csv("plots.csv")
- Once loaded you can display a data table by clicking on it in the “Environment” tab.
- This data set is a great example of a good tabular data structure
- One table per type of data
- Tables can be linked together to combine information.
- Each row contains a single record.
- A single observation or data point
- Each column or field contains a single attribute.
- A single type of information
- One table per type of data
The dplyr R package

- A modern library of R functions for data manipulation
- https://dplyr.tidyverse.org/
- “
dplyris a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges”. - When in doubt, use the cheat sheet:
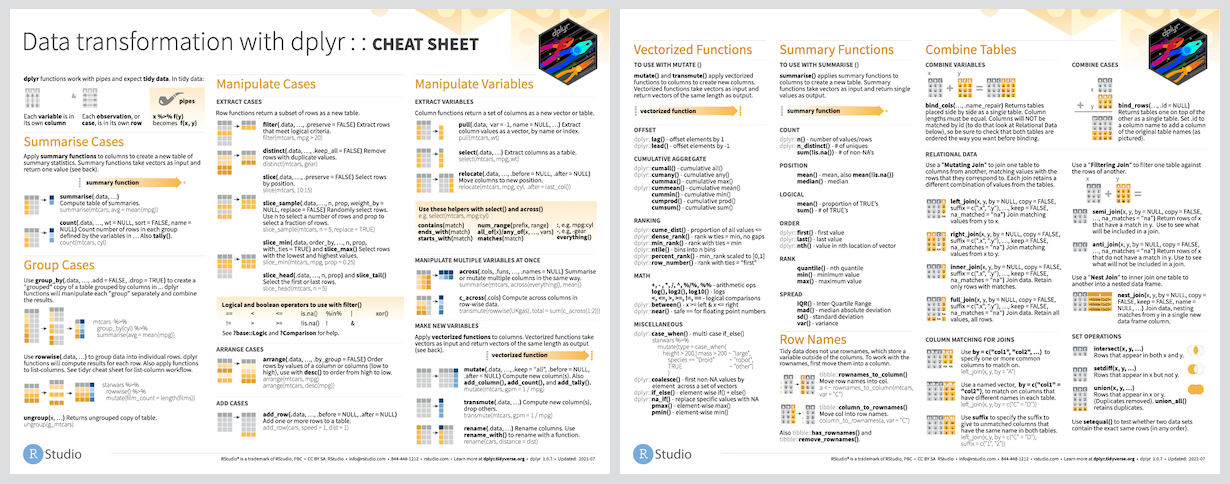
Installing vs loading packages
- Reminder: Packages are the main way that reusable code is shared in R
- Combination of code, data, and documentation
- R has a rich ecosystem of packages for data manipulation & analysis
- Download and install packages with the R console:
install.packages("dplyr")
- Even if we’ve installed a package it is NOT automatically available to do analysis with
- This is because sometimes different packages have functions with the same name
- So don’t want to have to worry about functions and packages we’ve installed every time we write a piece of code
- To use a package:
- Load all of the functions in the package with
library("dplyr") - Use the syntax
package_name::function_name
- Load all of the functions in the package with
Selecting columns
- Create a new data frame with your columns of interest, in any order you prefer using the function
select(). - For example, get a new data frame with the column “year”, “month” and “day” from the
surveysdata frame:
select(surveys, year, month, day)
- The function
select()does not modifiy the original object. Check this by comparing the output we just produced tosurveys:
head(surveys)
- Columns will follow the same order given in the function:
select(surveys, month, day, year)
Mutating data
- The function
mutate()creates new data columns based on data from existing columns - It allows to calculate new values using data from the original columns
- For example, the column
"hindfoot_length"stores measurements in millimeters (mm). To create a new column showing the length of the hindfoot in centimeters (cm), we have to divide the values in mm from the original column"hindfoot_length"by 10:
mutate(surveys, hindfoot_length_cm = hindfoot_length / 10)
- Similar to
select(), when we opensurveys, it does not contain a new column, because the functionmutate()does not modify the original object; it produces a new object with the new column. - To store the new data frame for later use we need to assign it to an object:
surveys_plus <- mutate(surveys,
hindfoot_length_cm = hindfoot_length / 10)
- Or we could overwrite the existing data frame if we don’t need it
surveys <- mutate(surveys,
hindfoot_length_cm = hindfoot_length / 10)
Arranging (sorting) data
- We can sort the data in the table using the function
arrange() - Let’s sort the surveys table by the values on the column
"weight":
arrange(surveys, weight)
- We can also sort by multiple columns, so if we wanted to sort first by
plot_idand then by date
arrange(surveys, plot_id, year, month, day)
- Similar to
select()andmutate(), the functionarrange()does not modifiy the original object. Check this by comparing the output we just produced tosurveys:
head(surveys)
- We can reverse the order of the sort by wrapping
"weight"in the functiondesc()(for “descending order”):
arrange(surveys, desc(weight))
Filtering values
- Use
filter()to keep only the rows that meet certain criteria. - We use relational or logical statements to evaluate and establish the criteria to filter on.
- Some examples of relational statements are:
1 == 1
1 == 2
1 != 2
1 > 2
1 > 1
1 >= 1
1 < 2
1 <= 2
"A" == "A"
"A" == "a"
"A" != "a"
- As with all other function that we have seen, the first argument specifies the data set we want to work with
- The second and following arguments are relational/conditional statements that specify the filtering criteria.
- For usage inside the function
filter(), relational/conditional statements are coded as:column namerelational/conditional statementvalue - For example, to filter the
surveysdata frame to only keep the data with the species id “DS”:- Type the name of the function and open parentheses,
filter() - The name of the data frame we want to filter,
surveys - The column the want to filter on,
"species_id" - The relational statement, which is
==for is equal to - And then the value we want to evaluate,
"DS" "DS"here is a string, not a variable or a column name, so we enclose it in quotation marks
- Type the name of the function and open parentheses,
filter(surveys, species_id == "DS")
- Like the examples above, we can have a relational statement that is not equal to using
!=. - For example, to filter the data for all species except “DS”:
filter(surveys, species_id != "DS")
- We can also filter on multiple criteria at once by adding more relational statements
- To indiciate that we want all the criteria to be TRUE, we have two ways:
- We can add more criteria for filtering by separating them with a comma
,.
- We can add more criteria for filtering by separating them with a comma
- For example, to get the data on species
"DS"for the year1995and above:
filter(surveys, species_id == "DS", year > 1995)
- Alternatively, we can use the ampersand
&symbol, which is called the AND operator:
filter(surveys, species_id == "DS" & year > 1995)
- Combining criteria with a comma
,or with the AND operator&indicates that we want all criteria to be met. - To indicate that not all statements have to be true (that is, one or more of the statements can be true), we combine statements using the vertical bar symbol
|, which is called the OR operator. - For example, to get data for all of the Dipodomys species, with different species id (“DS”, “DM”, and “DO”)
filter(surveys, species_id == "DS" | species_id == "DM" | species_id == "DO")
- The
&and|allow building filtering criteria that are as complex as needed.
Filtering missing values
NAis a special value in R that represents data that is missing and should be treated differently.- Think of
NAas “I do not know what’s there”. - One of the common tasks we use
filterfor is removing missing values from data - We know that the column
weighthas many missing values, so let’s filter out these values. - Based on what we learned before it’s natural to think that we do this by using the condition
weight != NA
filter(surveys, weight != NA)
- But that does not work. Let’s try some equivalences:
NA > 3 # is obviously NA because we don't know if the missing value is larger than 3 or not
NA == NA # the same with this, we have two missing values but the true values could be quite different, so the correct answer is "I don't know."
surveys$weight == NA
- Null values like
NAare special: they are incomparable. - We don’t want to accidentally say that two “missing” things are the same, because we don’t know if they are
- So we use special functions
- The function
is.na()checks if a value isNA
is.na(NA)
is.na(3)
is.na(surveys$weight)
filter(surveys, is.na(weight))
- To remove null values we combine the function
is.na()with the NOT operator!:
is.na(3)
!is.na(3)
filter(surveys, !is.na(weight))
- Keep the function
is.na()in mind. You will use it often to filter missing values in combination with other statements. - For example, we might want all of the data on a particular species that has weight data:
filter(surveys, species_id == "DS", !is.na(weight))
Solo In-class Exercise (30 min)
Exercise 1: Data manipulation
- Load
surveys.csvinto R usingread.csv(). - Use
select()to create a new data frame object calledsurveys1with just theyear,month,day, andspecies_idcolumns in that order. - Create a new data frame called
surveys2with theyear,species_id, and weight in kilograms of each individual, with no null weights. Usemutate(),select(), andfilter()with!is.na(). The weight in the table is given in grams so you will need to create a new column called “weight_kg” for weight in kilograms by dividing the weight column by 1000. - Use the
filter()function to get all of the rows in the data framesurveys2for the species ID “SH”.
Day 2: Pipes
The usual analysis workflow: intermediate variables and nesting functions
- We usually use intermediate variables when writing code
- We run one line of code for each action in sequential order
- Store the output in a variable
- Use that variable later in the code
- Repeat
- For example, to get the square root of the mean of a vector of numbers, we would run the
mean()function with a numeric vector as the input, assign it to a variable, and then run the functionsqrt()using that newly created object as input:
x = c(1, 2, 3)
mean_x <- mean(x)
sqrt_x <- sqrt(mean_x)
sqrt_x
- And, in nested form:
sqrt(mean(x = c(1,2,3)))
Pipes
- Intermediate variables and nested functions can get cumbersome (and confusing) if there are lots of steps.
%>%or|>(the pipe operator) takes the output of one command and passes it as value for the first argument of the next command.- The
|>pipe will work everywhere as long as you have a new enough version of R and RStudio %>%is the original pipe in R. It is called the “magrittr” pipe, and you have to load themagrittrpackage to use it (this gets loaded automatically bydplyr)magrittrhas some fancier functionality that may be useful in some cases- You can use any pipe you like.
- Shortcut to get the pipe:
Ctrl-shift-m.- You can change this to give the base R pipe: Tools -> Global Options -> Code -> Use native pipe operator
- To calculate the mean of a numeric vector, we can directly pipe the vector into the function
mean(), no need to create an object:c(1, 2, 3) %>% mean() - The numeric vector becomes the first argument in
mean - We can add other arguments to the functions as needed. For example, to remove missing values from the mean:
x = c(1, 2, 3, NA) mean(x, na.rm = TRUE) x %>% mean(na.rm = TRUE) - For example, taking the mean of
surveys$weightusing a pipe gives the same result as doing it using the classic sequential form:mean(surveys$weight, na.rm = TRUE) surveys$weight |> mean(na.rm = TRUE) - The usefulness (and clarity) of pipes becomes apparent when trying to run more complicated analysis, that require creating intermediate variables or nesting code:
c(1, 2, 3, NA) |> mean(na.rm = TRUE) |> sqrt()
Joint In-class exercise 3
- Write the necessary code using intermediate variables to manipulate the data as follows:
- (a) Use
mutate(),select(), andfilter()withis.na()to create a new data frame with theyear,species_id, and weight in kilograms of each individual, with no null weights. Create a new data object calledsurveys1. Remember: The weight in the table is given in grams so you will need to create a new column called “weight_kg” for weight in kilograms by dividing the weight column by 1000. - (b) Use
filter()withis.na()andselect()to get theyear,month,day, andspecies_idcolumns for all of the rows in the data frame wherespecies_idisSHand with no null weights. Create a new data object calledsurveys2.
- (a) Use
- Write the same code but using pipes (either
|>or%>%).
Solo In-class exercise 4
The following code is written using intermediate variables. It obtains the data for “DS” in the “species_id” column, sorted by year, with only the year and weight columns. Write the same code to get the same output but using pipes instead.
ds_data <- filter(surveys, species_id == "DS", !is.na(weight))
ds_data_by_year <- arrange(ds_data, year)
ds_weight_by_year <- select(ds_data_by_year, year, weight)
What if I want to pipe to an argument other than the first argument?
- To indicate the position of the piped argument we use the underscore symbol
_for the|>pipe and the dot symbol.for the%>%magrittr pipe. - This is used when the output goes to an argument other than the first argument of the function.
- For example, the function
lm()fits a linear model relationship between two variables. - This function takes the
formula =as the first argument and thedata =containing our two variables as the second argument. - For example, to evaluate if weight has decreased or increased with time, in sequential/nested form:
lm(weight ~ year, data = surveys) - And using pipes and the placeholder:
surveys |> lm(weight ~ year, data =_)
surveys %>%
lm(weight ~ year, data =.)
- We can get the summary of the fitted model with the function
summary(); in nested form:summary(lm(weight ~ year, data = surveys))
Solo In-class exercise 5
Use pipes to evaluate and summarize the relationship between weight and year for the species “DS”. Make sure that you filter for missing values in weight. The code in sequential form would look like the following:
surveys_DS <- filter(surveys, species_id == "DS", !is.na(weight))
surveys_DS_lm <- lm(weight ~ year, data = surveys_DS)
summary(surveys_DS_lm)
Data grouping (also called data agreggation)
Basic grouping
- The function
group_by()combines rows into groups based on ONE or MORE columns. group_by()function has two arguments: 1) data to work on; 2) name of column (or columns) to group by- For example, to group
surveysbyyear:
group_by(surveys, year)
- The output is a
tibble, a different looking kind ofdata.frame.- The
tibblecontains information on the data source, the groupings, and the data type.
- The
- We can also group by multiple columns.
- For example, group by “plot id” and by “year”:
group_by(surveys, plot_id, year)
Summarizing data from groupings
- After grouping a data frame we can use the function
summarize()to analyze each group. - The function
n()counts the number of rows for each grouping. It is a special function that only works withindplyrfunctions. - The
summarize()function takes as first argument a grouped table, an output ofgroup_by() - Then, it takes one additional argument for each new column with summarized data you want to add.
- The format of this arguments is
New column name, followed by the equal sign=, finished by the calculation that we want to perform for each group - For example, to get the abundance of individual pers year, we will use the function
n()
surveys_by_year <- group_by(surveys, year)
year_counts <- summarize(surveys_by_year, abundance = n())
- Count the number of individuals in each plot in each year
surveys_by_plot_year <- group_by(surveys, plot_id, year)
plot_year_counts <- summarize(surveys_by_plot_year, abundance = n())
- Just like with other
dplyrfunctions we could write this using pipes instead
plot_year_counts <- surveys |>
group_by(plot_id, year) |>
summarize(abundance = n())
- We can use any function that returns a single value from a vector; e.g.,
mean(),max(),min(),sum() - For example, we can calculate the number of individuals in each plot-year group and their average weight
surveys |>
group_by(plot_id, year) |>
summarize(abundance = n(), avg_weight = mean(weight))
- We get all
NAin the “avg_weight” column :/ - Remember:
mean()(and max, min and sum) returnsNAwhen the input vector has missing values (NAs) - To fix this we use the argument
na.rm = TRUE:mean(surveys$weight, na.rm = TRUE) - Now, applied to the pipe:
surveys |> group_by(plot_id, year) |> summarize(abundance = n(), avg_weight = mean(weight, na.rm = TRUE)) - We still get missing valies (in the form of
NaN) because there are species that have never been weighed! - We can
filter()this using!is.na()
surveys |>
group_by(plot_id, year) |>
summarize(abundance = n(),
avg_weight = mean(weight, na.rm = TRUE)) |>
filter(!is.na(avg_weight))
Solo In-class Exercise 6
Exercise 3: Data agreggation
- Use the
group_by()andsummarize()functions to get a count of the number of individuals in each species ID. - Use the
group_by()andsummarize()functions to get a count of the number of individuals in each species ID in each year. - Use the
filter(),group_by(), andsummarize()functions to get the mean mass of speciesDOin each year.
