Day 1: Joining data tables
Learning Objectives
This week, students will learn to:
- Explain the importance of joining multiple data tables.
- Use the
dplyrfunctions that join data tables.- Understand why data is dropped when joining tables
- Use pipes to join more than two data tables
- Use the
%in%operator to find matching column names in two data tablesPractice Objectives
This week, students will practice:
- Use of relational and logical statements to filter data tables
- Handling missing values with
is.na()andna.rm =- pipeline placeholders
Non Objectives
which()match()
Homework and class review (15 min)
- review of homework
select(),mutate(),filter(),arrange()do not change the original data!- We use relational/logical statements as filtering criteria in the function
filter() - Some examples of simple relational statements are:
1 == 1 1 == 2 1 != 2 1 > 2 1 > 1 1 >= 1 1 < 2 1 <= 2 "A" == "A" "A" == "a" "A" != "a" - We use two general types of logical statements: AND, and OR
- Some examples of logical statements:
1 == 1 & 1 == 2 1 == 1 | 1 == 2 - relational operations with
NANA > 3 # is obviously NA because we don't know if the missing value is larger than 3 or not NA == NA # the same with this, we have two missing values but the true values could be quite different, so the correct answer is "I don't know." surveys$weight == NA - That’s why we have
is.na(), a special function to detectNAvalues. - Also, the argument
rm.na = TRUEdetects and removes missing values in functionsmean(),max(),sum(),min(). - The classic coding workflow: intermediate variables and nesting functions
- The new (in R), cleaner alternative: the coding pipeline
- pipes work by taking the output from a function and giving it to a second function without creating intermediate objects or variables
- by default, they feed the output to the first argument!
Setup your RStudio project (15 min)
- Remember! Working in projects is considered a best practice in data science.
- Go ahead and open your RStudio project for the class. I called mine fall-2022.
- We will keep on working on:
- the rmarkdown file
"small-mammals.Rmd", and - the Portal Project Teaching Database.
- the rmarkdown file
- If you have not downloaded the following CSV files, do so by clicking on their names:
- surveys.csv - main table, one row for each rodent captured, date on date, location, species ID, sex, and weight in grams and hindfoot length in millimeters.
- species.csv - latin species names for each species ID + general taxon
- plots.csv - information on the experimental manipulations at the site
- Make sure that the three CSV files are:
- saved into your data-raw folder.
- loaded into
Rusing the functionread.csv()from the packageutils.- Remember to use relative paths!
surveys <- read.csv("surveys.csv") species <- read.csv("species.csv") plots <- read.csv("plots.csv")
- Remember to use relative paths!
- Once loaded, we can display the files as a data table by clicking on their name in the “Environment” tab.
- This data set is a great example of a good tabular data structure
- One table per type of data
- Tables can be linked together to combine information.
- Each row contains a single record.
- A single observation or data point
- Each column or field contains a single attribute.
- A single type of information
- One table per type of data
Why do we need to join data tables? (5 min)
- One of the best practices in data science is to structure data from the same project/experiment in multiple tables:
- It is recommended to have one main table (the
surveystable in our case) and multiple supplementary tables that provide additional details.
- It is recommended to have one main table (the
- The practical reasons why data scientists do this are:
- it avoids redundant information (like listing the full taxonomy for every individual of a species),
- it makes storage more efficient (smaller files),
- it makes data processing more efficient (such as changing data in one place, not hundreds of places),
- it makes tables more readable, as they contain a single kind of information.
- So, having data in multiple tables means that we often need to join them to perform an analysis.
Joining two data Tables (5 min)
- To combine two tables based on the values from a shared column, we use the
dplyrfunctionsinner_join()left_join()right_join()full_join()
- These functions take at least three arguments:
- The first two arguments specify the two tables that we want to join.
- The third argument
by =specifies the name of the shared column as a character string (enclosed in quotations" ") - For example, to join the tables
surveysandspeciesby their only shared column"species_id"into a newcombinedtable:inner_join(surveys, species, by = "species_id") - The same, but with a pipe:
surveys |> inner_join(species, by = "species_id")
Exercise 1 (10 min)
Do the following calculations using a single pipe of code (no nested nor intermediate variables):
- Use
inner_join()andfilter()to get a data frame with the information from thesurveysandplotstables where the"plot_type"is"Control".
Check dropped data
- The function
nrow()let us see the row numbers from the original tables and the joined tables:nrow(surveys) nrow(combined) - We can test if the row number has changed:
nrow(surveys) == nrow(combined) - inner joins: only keep information from both tables when both tables have a matching value in the join column
- Rows with
"species_id"values from the first table that are not in the second table (and visceversa) are dropped: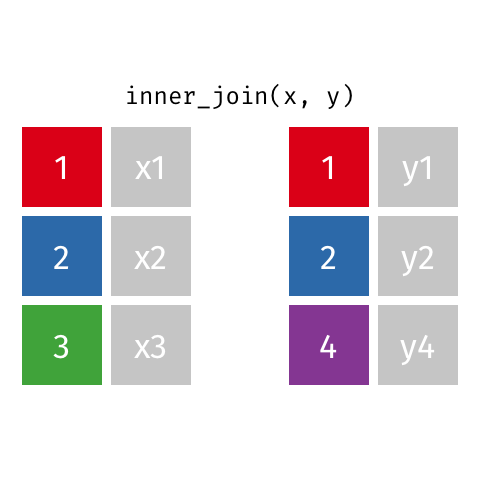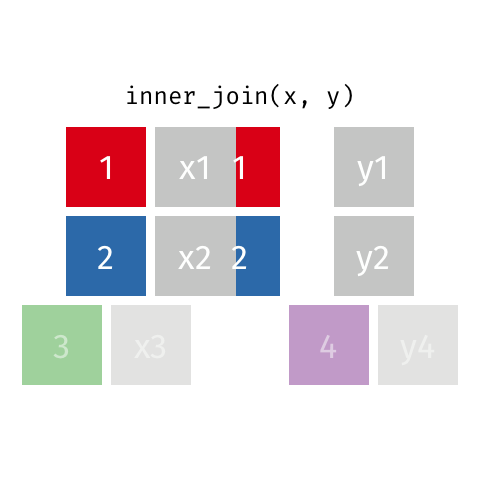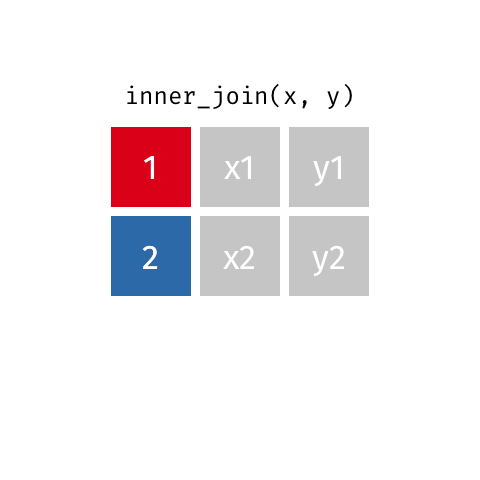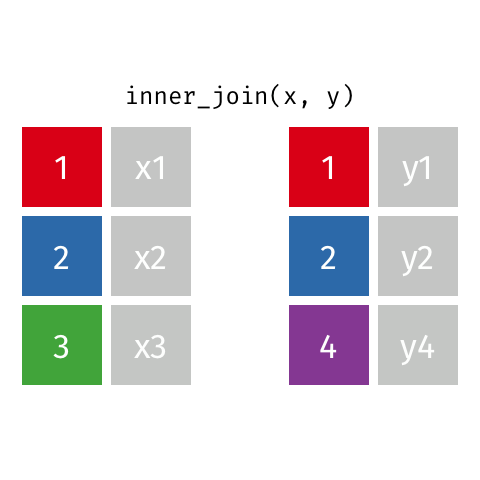
- Rows with
- left joins: The function
left_join()keeps all values from the left table (the first table given in the function). - right joins:
right_join()keeps all values from the right table (the second table given in the function). - full joins: keep all information from both tables.
Finding shared column names (colnames()) between tables (5 min)
- How do we find shared column names to join our tables??
- We can visually search for shared column names between two tables:
- We can open each table from the Environment tab, or with the function
View() - We can display the
colnames()of each table individually 2. Using code! - The function
intersect():intersect(colnames(surveys), colnames(species))
Exercise 1 (10 min)
- Find the column name that is shared between the
plotstable and thesurveystable. Use that column name for the next question. - Do the following using a single pipe of code (no nested code nor intermediate variables):
- Use function
inner_join()andfilter()to get a data frame with the information from thesurveysandplotstables where the"plot_type"is"Control".
- Use function
Joining two or more data Tables (5 min)
- There is no special function to join more than two data tables.
- We use the
_join()functions, incrementally:- Start by joining two tables
- Then, join the resulting table to a third table, and so on.
- For example, for the Portal data set, we can start by joining the surveys and the species tables together, and then combining the resulting table with the plots table:
combined <- inner_join(surveys, species, by = "species_id") combined_final <- inner_join(combined, plots, by = "plot_id")
- Now, how do we do this using a pipe?
combined <- surveys |> inner_join(species, by = "species_id") |> inner_join(plots, by = "plot_id")
Exercise 2 (15 min)
We want to do an analysis comparing the size of individuals on the "Control" plots to the "Long-term Krat Exclosures".
- Create a data frame with the
"year","genus","species","weight"and"plot_type"for all cases where the - plot type is either
"Control"or"Long-term Krat Exclosure". Pay attention to typos in lower case and upper case values. - Only include cases where the column
"taxa"is"Rodent". - Remove any records where the
"weight"is missing.
Start with the Homework
Exercises 3 and 4 of Joining data tables practice.
Day 2: Joining data vectors
Learning Objectives
This week, students will learn to:
-
- - -
Practice Objectives
This week, students will practice:
-
Non Objectives
-
Setup Your RStudio Project
- Remember! Working in projects is considered a best practice in data science.
- Go ahead and open your RStudio project for the class. I called mine fall-2022.
- We will keep on working on:
- the rmarkdown file
"small-mammals.Rmd", and - the Portal Project Teaching Database.
- the rmarkdown file
- If you have not downloaded the following CSV files, do so by clicking on their names:
- surveys.csv - main table, one row for each rodent captured, date on date, location, species ID, sex, and weight in grams and hindfoot length in millimeters.
A Relationship Between Data Frames and Vectors
- R is a programming language for data anlysis
- It stores information using data structures
- We’ve learned about two general ways to store data, vectors and data frames
- What are vectors: Vectors store a single set of values with the same type
- What are data frames: Data frames store multiple sets of values, one in each column, that can have different types
- These two ways of storing data are related to one another
- Actually, all data structures in R are related to each other!
- A data frame is a bunch of equal length vectors that are grouped together
- So, we can extract vectors from data frames and we can also make data frames from vectors
Creating vectors
- Examples of numeric vectors:
- with a single number
a_number <- 1 - with two or more numbers, we can use the concatenate function
c()c(1, 2, 3) # in order - We can use the colon
:operator to create sequences of numbers1:3 - With the
c()function we can add numbers in any order we wantc(10, 1, 8) # random order - But with
:we can create sequences as long as we want to with just a few key strokes:1:10 1:100 1:4589567 - The function
seq()creates sequences with any step we specify (not only 1 as with:)seq(from = 1, to = 100, by = 2) seq(from = 1, to = 100, by = 0.5) - We can start numeric sequences at any number, in reverse order, and using negative numbers,
- with the
:operator:15:20 100:50 -100:50 5:-5 - and with
seq()(pay attention to the sign of the step (by =argument))seq(15, 20) seq(100, 50, -2) seq(-100, 50, 2)
- with the
- with a single number
- Examples of logical vectors
abc
Creating data frames from Vectors
- The
data.frame()function joins vectors into a single data frame - Each argument we provide will be a column in the data frame (just like in
mutate()andsummarize()!) - The arguments are taken as follows:
- The name of the column we want in the data frame,
- an equal sign
=, and - the vector whose values we want in that column.
- So we give it the arguments
sites =, anddensity =density_data <- data.frame(sites = c("a", "a", "b", "c"), density = c(2.8, 3.2, 1.5, 3.8)) - If we look in the Global Environment tab we can see that there is a new data frame called
density_data, and that it has our two vectors as columns. - We could also make this data frame using the vectors that are already stored in variables:
sites <- c("a", "a", "b", "c") density <- c(2.8, 3.2, 1.5, 3.8) density_data <- data.frame(sites = sites, density = density) - We can also add columns to the data from that only include a single value without first creating a vector
- We do this by providing a name for the new column, an equals sign, and the value that we want to occur in every row
- For example, if all of this data was collected in the same year and we wanted to add that year as a column in our data frame we could do it like this:
density_data_year <- data.frame(year = 2000, sites = sites, density = density)
year =sets the name of the column in the data frame- And 2000 is that value that will occur on every row of that column
- If we run this and look at the
density_data_yeardata frame we’ll see that it includes the year column with the value2000in every row
Joint in-class exercise
You have data on the length, width, and height of 10 individuals of the yew Taxus baccata stored in the following vectors:
length <- c(2.2, 2.1, 2.7, 3.0, 3.1, 2.5, 1.9, 1.1, 3.5, 2.9)
width <- c(1.3, 2.2, 1.5, 4.5, 3.1, NA, 1.8, 0.5, 2.0, 2.7)
height <- c(9.6, 7.6, 2.2, 1.5, 4.0, 3.0, 4.5, 2.3, 7.5, 3.2)
- Make a data frame that contains these three vectors as columns along with a
"genus"column containing the genus name Taxus on all rows and a"species"column containing the species epithet baccata on all rows.
Extracting values from vectors and data frames
Extracting vectors from data frames
- There are several ways to extract a vector from a data frame
- Let’s look at these using the Portal data
- We’ll start by loading the
surveystable into R
surveys <- read.csv("surveys.csv")
- One common approach to extracting a column into a vector is to use the
[] - Remember that
[]also mean “give me a piece of something” - Let’s get the
species_idcolumn "species_id"has to be in quotes because we we aren’t usingdplyr
surveys["species_id"]
- This actually returns a one column data frame, not a vector
- To extract a single column as a vector we use two sets of
[] - Think of the second set of
[]as getting the single vector from inside the one column data frame
surveys[["species_id"]]
- We can also do this using
$ - The
$in R is short hand for[[]]in cases where the piece we want to get has a name - So, we start with the object we want a part of, our
surveysdata frame - Then the
$with no spaces around it - and then the name of the
species_idcolumn (without quotes, just to be confusing)
surveys$species_id
Exercise
Using the Portal data surveys table (download a copy if it’s not in your working directory):
- Use
$to extract theweightcolumn into a vector calledsurveys_weight - Use
[]to extract themonthcolumn into a vector calledsurveys_month - Extract the
hindfoot_lengthcolumn into a vector and calculate the mean hindfoot length ignoring missing values.
Extracting Values from Vectors
letters[10] # indexing the 10th letter of the alphabet
letters[1:3] # getting the first three letters
abc <- letters[c(1,2,3)] # creating a vector of the first three letters of the alphabet
letters[3:1] #
letters[-1]
letters[-1:5]
Overwriting values in vectors and data frames
Summary
- A data frame is a set of equal length vectors
- We can extract a column of a data frame into a vector using either
$or two sets of square brackets[[]] - We can combine vectors into data frames using the
data.frame()function, which takes a series of arguments, one vector for each column we want to create in the data frame.
