Day 1: Functions in R
Learning Objectives
This week, students will:
- learn the parts of a function
Practice Objectives
- Reading and writing tables
- Reading and plotting phylogenies
Setup your RStudio project (5 min)
- 🎗️ Structuring your files into a project is a best practice for good data science!
- Open your RStudio project for the class; I called mine “spring2023”.
- Open a new file, name it “functions.Rmd”, and save it to your “documents” folder.
Writing functions (10 min)
- In this lesson we will use the concept of “mass” to learn how to create functions.
- Mass describes the density and type of atoms in any given object.
- The SI unit of mass is the kilogram (kg), though mass can also be measured in grams (gr), ounces (oz), and pounds (lb).
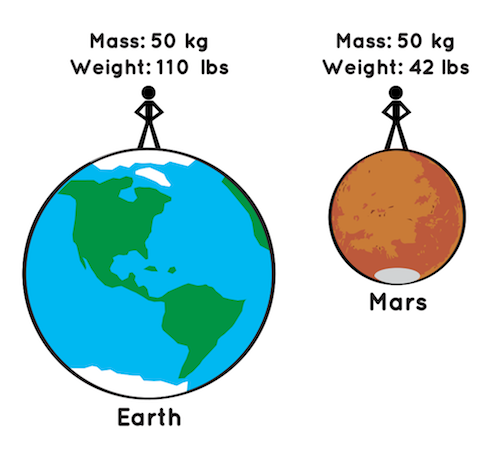
- Reminder - Mass is not the same as weight:
- Weight is the measure of the force of gravity on an object
- It is hence a relative measure
- Mass is an absolute measure
- The mass of an object will never change, but the weight of an item can change based on its location.
- Formulas to convert between different units of mass:
| Pounds to kilograms | Kilograms to pounds |
|---|---|
 |
 |
 |
 |
Exercise 1: Converting between units of mass
Using and modifying functions (20 min)
- The length of an organism is strongly correlated with its body mass.
- This allometric relationship takes the form of mass equals to parameter “a” multiplied by “length” elevated to the power of parameter “b”:
mass = a * length^b
- Parameters
aandbvary among biological groups. - Scientists use this formula to estimate the mass of organisms for which we only have length measurememts. For example:
- trees, we cannot weigh them unless we unroot them,
- extinct creatures such as dinosaurs, as we cannot get the living weight of something that is fossilized.
- The following function uses the formula
mass = a * length^bto estimate the mass of an organisms belonging to Theropoda dinosaurs in kg, based on its length in meters and the set of parameter valuesaandbthat have been estimated for that biological group,a = 0.73andb = 3.63, by Seebacher (2001).- Take 3 min to type the following function in:
get_mass_from_length_theropoda <- function(length){ mass <- 0.73 * length ^ 3.63 return(mass) }
- Take 3 min to type the following function in:
Exercise 2: The weight of dinosaurs
Why do we use functions in programming? (15 min)
Discussion in small groups (5 min)
-
Discuss in small groups: Based on your experience with coding so far and after watching the introductory videos, how does creating your own functions would improve your coding workflow?
-
Individually, work on your Rmd file:
- Write a second level subtitle for the introductory section.
- List and justify three reasons why using functions is useful in data science.
Discussion in full (10 min)
-
How does creating your own functions will improve your code?
- Makes code more understandable:
- Code is shorter, easier to remember
- Code is more organized for you and others, so it is easier to read
- Code is grouped conceptually, easier to understand
- Code is more manageable, invites you to be intentional on the code that you are writing
- Allows to be more in control of the outputs
- Makes code reusable:
- It allows reusing code for other parts of a project or a future project
- It is less error prone than copy-pasting code
- If it occurs in more than one place, it will eventually be wrong somewhere.
- It is more efficient than copy-pasting code
- Functions are written to be reusable.
Defining argument values in a function (20 min)
- In the previous exercise, you created the function
get_mass_from_length()which is a more flexible form ofget_mass_from_length_theropoda(), by allowingaandbto be passed as arguments. - Still, for some organisms we don’t have specific values of
aandb. In this case, we have to use values ofaandbthat can be applied generally. - We can give this general values as default values for some or all arguments in any function that we are creating.
Exercise 3: The general form of a formula
Combining functions (20 min)
- The metric system is the standard approach used in scientific practice.
- To communicate scientific results to a broader audience, it might be more impactful to use different units (at least in some countries).
Exercise 4: Facilitating impactful science
Homework:
Instructions
- Open a new file called “functions-after.Rmd” that is saved in your “documents” folder.
- Write the code to solve the following exercises in R chunks.
- Add comments to each line of code explaining with your own words what the code is doing.
- Once you are finished, knit to PDF.
- Git add, commit and push the new files (PDF and Rmd) to your remote repository.
Exercise 5: Creating a function for the Portal data set
Day 2: Making choices inside R functions
Review from last class (10 min)
- Exercise 4 from last class: Combining functions
- Combining functions in a nested way
- Calling functions inside functions
- Homework
Setup your RStudio project (5 min)
- 🎗️ Structuring your files into a project is a best practice for good data science!
- Open your RStudio project for the class; I called mine “fall-2022”.
- Open a new file, name it “choices.Rmd”, and save it to your “documents” folder.
Review: Logical and conditional statements (15 min)
- Statements that return logical values (
TRUEorFALSE)- logical operators
- conditional operators
- logical functions
Exercise 6: Practice logical and conditional statements
Basic if statement (10 min)
- A simple
ifstatement allows us to choose between a single option and its alternative.
Exercise 7: Handling one choice
The if/else statement (10 min)
- An
elsestatement allows us to choose between two options and its alternative.
Exercise 8: Handling 2 choices
The if/else if/else statement (10 min)
- An
else ifstatement allows us to choose between three or more options and an alternative.
Exercise 9: Handling 3 choices or more
Using conditions inside functions (30 min)
- We can use conditions inside functions
- Conditions alter the behavior of a functions
- Conditions give us more control on the behavior of a function
Exercise 10: Value of y by age class (10 min)
- Conditions allow us to be even more efficient in reusing code
- For example, we can consolidate all the functions we created to get the mass of a dinosaur into a single one.
Exercise 11: Mass estimates by biological group (20 min)
Homework:
Instructions
- Open a new file called “choices-after.Rmd” that is saved in your “documents” folder.
- Write the code to solve the following exercises in R chunks.
- Add comments to each line of code explaining with your own words what the code is doing.
- Once you are finished, knit to PDF.
- Git add, commit and push the new files (PDF and Rmd) to your remote repository.
