Day 1: Joining data tables
Learning Objectives
This week, students will learn to: - Explain the importance of joining multiple data tables. - Use the `dplyr` functions that join data tables. - Understand why data is dropped when joining tables - Use pipes to join more than two data tables - Use the ``%in%`` operator to find matching column names in two data tables **Practice Objectives** This week, students will practice: - Use of relational and logical statements to filter data tables - Handling missing values with `is.na()` and `na.rm =` - pipeline placeholders **Non Objectives** - `which()` - `match()`Quizz (10 min)
Answer the following questions on the etherpad:
- What type of output does the
summarize()function fromdplyrgive, a table or a vector? - When using the
summarize()function, does the output table has the same columns as the input data table or different? - Mention 3 functions from the
dplyrpackage that return tables with (at least) the same columns from the input table?
Setup your RStudio project (15 min)
- Remember! Working in projects is considered a best practice in data science.
- Go ahead and open your RStudio project for the class. I called mine sprng2023.
- We will keep on working on the data set from
- If you have not downloaded the following CSV files, do so by clicking on their names:
- surveys.csv - main table, one row for each rodent captured, date on date, location, species ID, sex, and weight in grams and hindfoot length in millimeters.
- species.csv - latin species names for each species ID + general taxon
- plots.csv - information on the experimental manipulations at the site
- Make sure that the three CSV files are saved into your data-raw folder.
- Create a new Rmd file named
"joining-tables.Rmd"and save it in yourDocumentsfolder. - Load the 3 tables into
Rusing the functionread.csv()from the packageutils.- Remember to use relative paths!
surveys <- read.csv("surveys.csv") species <- read.csv("species.csv") plots <- read.csv("plots.csv")
- Remember to use relative paths!
- Once loaded, we can display the files as a data table by clicking on their name in the “Environment” tab.
- This data set is a great example of a good tabular data structure
- One table per type of data
- Tables can be linked together to combine information.
- Each row contains a single record.
- A single observation or data point
- Each column or field contains a single attribute.
- A single type of information
- One table per type of data
Why do we need to join data tables? (5 min)
- One of the best practices in data science is to structure data from the same project/experiment in multiple tables:
- It is recommended to have one main table (the
surveystable in our case) and multiple supplementary tables that provide additional details.
- It is recommended to have one main table (the
- The practical reasons why data scientists do this are:
- it avoids redundant information (like listing the full taxonomy for every individual of a species),
- it makes storage more efficient (smaller files),
- it makes data processing more efficient (such as changing data in one place, not hundreds of places),
- it makes tables more readable, as they contain a single kind of information.
- So, having data in multiple tables means that we often need to join them to perform an analysis.
Joining two data Tables (5 min)
- To combine two tables based on the values from a shared column, we use the
dplyrfunctionsinner_join()left_join()right_join()full_join()
- These functions take at least three arguments:
- The first two arguments specify the two tables that we want to join.
- The third argument
by =specifies the name of the shared column as a character string (enclosed in quotations" ") - For example, to join the tables
surveysandspeciesby their only shared column"species_id"into a newcombinedtable:inner_join(surveys, species, by = "species_id") - The same, but with a pipe:
surveys |> inner_join(species, by = "species_id")
Exercise 1 (5 min)
Do the following calculations using a single pipe of code (no nested nor intermediate variables):
- Use
inner_join()andfilter()to get a data frame with the information from thesurveysandplotstables where the"plot_type"is"Control".
Check dropped data
- The function
nrow()let us see the row numbers from the original tables and the joined tables:nrow(surveys) nrow(combined) - We can test if the row number has changed:
nrow(surveys) == nrow(combined) - inner joins: only keep information from both tables when both tables have a matching value in the join column
- Rows with
"species_id"values from the first table that are not in the second table (and visceversa) are dropped: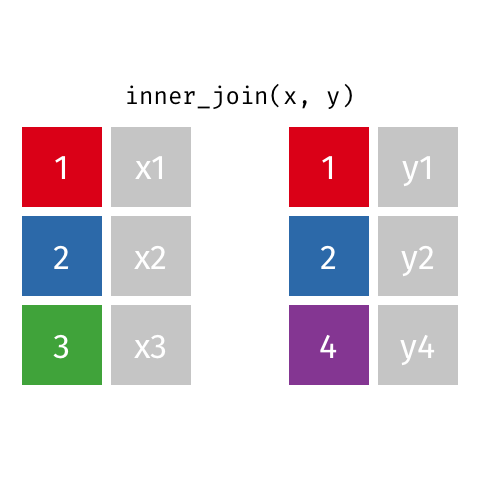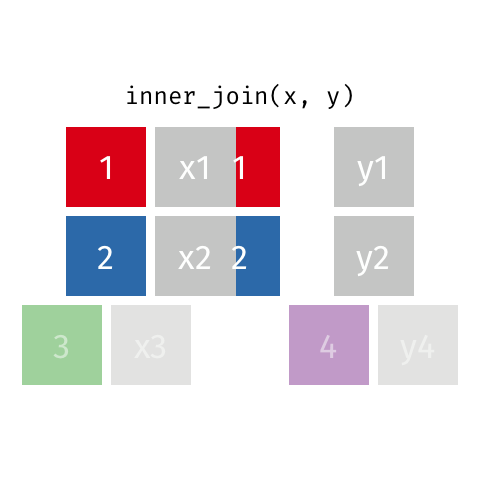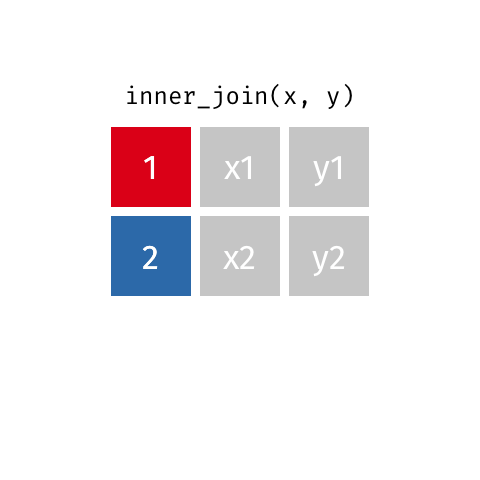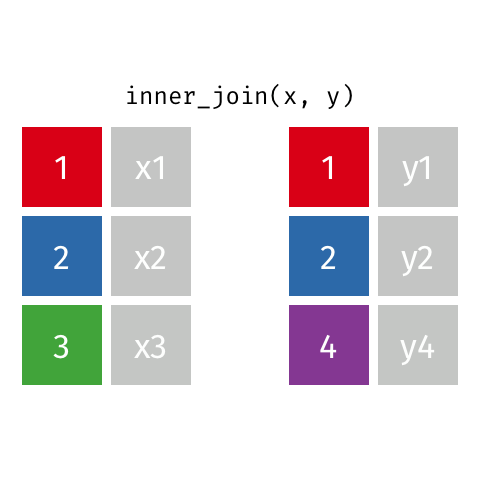
- Rows with
- left joins: The function
left_join()keeps all values from the left table (the first table given in the function). - right joins:
right_join()keeps all values from the right table (the second table given in the function). - full joins: keep all information from both tables.
Finding shared column names (colnames()) between tables (5 min)
- How do we find shared column names to join our tables??
- We can visually search for shared column names between two tables:
- We can open each table from the Environment tab, or with the function
View() - We can display the
colnames()of each table individually 2. Using code! - The function
intersect():intersect(colnames(surveys), colnames(species))
Exercise 2 (5 min)
- Use
intersect()to display the column name that is shared between theplotstable and thesurveystable. Use that column name for the next question. - Do the following using a single pipe of code (no nested code nor intermediate variables):
- Use function
left_join()andfilter()to get a data frame with the information from thesurveysandplotstables where the"plot_type"is"Rodent Exclosure".
- Use function
Joining two or more data Tables (5 min)
- There is no special function to join more than two data tables.
- We use the
_join()functions, incrementally:- Start by joining two tables
- Then, join the resulting table to a third table, and so on.
- For example, for the Portal data set, we can start by joining the surveys and the species tables together, and then combining the resulting table with the plots table:
combined <- inner_join(surveys, species, by = "species_id") combined_final <- inner_join(combined, plots, by = "plot_id")
- Now, how do we do this using a pipe?
combined <- surveys |> inner_join(species, by = "species_id") |> inner_join(plots, by = "plot_id")
Exercise 3 (10 min)
We want to do an analysis comparing the size of individuals on the "Control" plots to the "Long-term Krat Exclosures".
- Create a data frame with the
"year","genus","species","weight"and"plot_type"for all cases where the - plot type is either
"Control"or"Long-term Krat Exclosure". Pay attention to typos in lower case and upper case values. - Only include cases where the column
"taxa"is"Rodent". - Remove any records where the
"weight"is missing.
A minute feedback for class 17
- Please provide some quick feedback for this session here.
Homework
- Do at least one exercise from Joining data tables practice.
Day 2: Joining data vectors
Setup Your RStudio Project
- Remember! Working in projects is considered a best practice in data science.
- Go ahead and open your RStudio project for the class. I called mine spring2023.
- Create a new Rmd file and name it
"joining-vectors.Rmd", and - If you have not downloaded the following CSV file, do so by clicking on its name:
- surveys.csv - main table, one row for each rodent captured, date on date, location, species ID, sex, and weight in grams and hindfoot length in millimeters.
A Relationship Between Data Frames and Vectors
- R is a programming language for data anlysis
- It stores information using data structures
- We’ve learned about two general ways to store data, vectors and data frames
- What are vectors: Vectors store a single set of values with the same type
- What are data frames: Data frames store multiple sets of values, one in each column, that can have different types
- These two ways of storing data are related to one another
- Actually, all data structures in R are related to each other!
- A data frame is a bunch of equal length vectors that are grouped together
- So, we can extract vectors from data frames and we can also make data frames from vectors
Creating vectors
Review
- Examples of numeric vectors: - with a single number ```r a_number <- 1 ``` - with two or more numbers, we can use the _concatenate_ function `c()` ```r c(1, 2, 3) # in order ``` - We can use the _colon_ `:` operator to create sequences of numbers ```r 1:3 ``` - With the `c()` function we can add numbers in any order we want ```r c(10, 1, 8) # random order ``` - But with `:` we can create sequences as long as we want to with just a few key strokes: ```r 1:10 1:100 1:4589567 ``` - The function `seq()` creates sequences with any step we specify (not only 1 as with `:`) ```r seq(from = 1, to = 100, by = 2) seq(from = 1, to = 100, by = 0.5) ``` - We can start numeric sequences at any number, in reverse order, and using negative numbers, - with the `:` operator: ```r 15:20 100:50 -100:50 5:-5 ``` - and with `seq()` (pay attention to the sign of the step (`by = ` argument)) ```r seq(15, 20) seq(100, 50, -2) seq(-100, 50, 2) ```
### Creating data frames from Vectors
* The `data.frame()` function joins vectors into a single data frame
* Each argument we provide will be a column in the data frame (just like in `mutate()` and `summarize()`!)
* The arguments are taken as follows:
- The name of the column we want in the data frame,
- an equal sign `=`, and
- the vector whose values we want in that column.
* So we give it the arguments `sites = `, and `density = `
```r
density_data <- data.frame(sites = c("a", "a", "b", "c"), density = c(2.8, 3.2, 1.5, 3.8))
```
* If we look in the **Global Environment** tab we can see that there is a new data frame called `density_data`, and that it has our two vectors as columns.
* We could also make this data frame using the vectors that are already stored in variables:
```r
sites <- c("a", "a", "b", "c")
density <- c(2.8, 3.2, 1.5, 3.8)
density_data <- data.frame(sites = sites, density = density)
```
* We can also add columns to the data from that only include a single value without first creating a vector
* We do this by providing a name for the new column, an equals sign, and the value that we want to occur in every row
* For example, if all of this data was collected in the same year and we wanted to add that year as a column in our data frame we could do it like this:
```r
density_data_year <- data.frame(year = 2000, sites = sites, density = density)
```
* `year =` sets the name of the column in the data frame
* And 2000 is that value that will occur on every row of that column
* If we run this and look at the `density_data_year` data frame we'll see that it includes the year column with the value `2000` in every row
### Exercise 6
You have data on the length, width, and height of 10 individuals of the yew *Taxus baccata* stored in the following vectors:
```r
length <- c(2.2, 2.1, 2.7, 3.0, 3.1, 2.5, 1.9, 1.1, 3.5, 2.9)
width <- c(1.3, 2.2, 1.5, 4.5, 3.1, NA, 1.8, 0.5, 2.0, 2.7)
height <- c(9.6, 7.6, 2.2, 1.5, 4.0, 3.0, 4.5, 2.3, 7.5, 3.2)
```
- Make a data frame that contains these three vectors as columns along with a `"genus"` column containing the genus name *Taxus* on all rows and a `"species"` column containing the species epithet *baccata* on all rows.
---
---
### Extracting values from vectors and data frames
#### Extracting vectors from data frames
* There are several ways to extract a vector from a data frame
* Let's look at these using the Portal data
* We'll start by loading the `surveys` table into R
```r
surveys <- read.csv("surveys.csv")
```
* One common approach to extracting a column into a vector is to use the `[]`
* Remember that `[]` also mean "give me a piece of something"
* Let's get the `species_id` column
* `"species_id"` has to be in quotes because we we aren't using `dplyr`
```r
surveys["species_id"]
```
* This actually returns a one column data frame, not a vector
* To extract a single column as a vector we use two sets of `[]`
* Think of the second set of `[]` as getting the single vector from inside the one column data frame
```r
surveys[["species_id"]]
```
* We can also do this using `$`
* The `$` in R is short hand for `[[]]` in cases where the piece we want to get has a name
* So, we start with the object we want a part of, our `surveys` data frame
* Then the `$` with no spaces around it
* and then the name of the `species_id` column (without quotes, just to be confusing)
```r
surveys$species_id
```
### Exercise 7
Using the Portal data `surveys` table ([download a copy](https://ndownloader.figshare.com/files/2292172) if it's not in your working directory):
1. Use `$` to extract the `weight` column into a vector called `surveys_weight`
2. Use `[]` to extract the `month` column into a vector called `surveys_month`
3. Extract the `hindfoot_length` column into a vector and calculate the mean hindfoot length ignoring missing values.
### Extracting Values from Vectors
```r
letters[10] # indexing the 10th letter of the alphabet
letters[1:3] # getting the first three letters
abc <- letters[c(1,2,3)] # creating a vector of the first three letters of the alphabet
letters[3:1] #
letters[-1]
letters[-1:5]
```
### Overwriting values in vectors and data frames
### Summary
* A data frame is a set of equal length vectors
* We can extract a column of a data frame into a vector using either `$` or two sets of square brackets `[[]]`
* We can combine vectors into data frames using the `data.frame()` function, which takes a series of arguments, one vector for each column we want to create in the data frame.
**A minute feedback for class 18**
- Please provide some quick feedback for this session [here](https://docs.google.com/forms/d/e/1FAIpQLSfkCzaYH7qgLgeF9b9KLrCZJSfVmu0AYGLZo2MINBSN04B2tA/viewform).
### Homework
- Do 2 exercises from [Shrub volume data set - part 2](/spring2023-data-science/assignments/assignment_data_tables_2/).
