6. Getting branch length information (proportional to time) for you taxa
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I find supporting trees that include branch lengths?
How do I subset them to include just the taxa I am interested in?
Objectives
Learn about the
opentree_chronogramsobject from datelife.Get source chronograms from the
opentree_chronogramsobject for a set of taxa.
What if I want to search the OTOL database (phylesystem) for studies and trees matching some criteria?
The rotl package has functions that allow getting a list of studies or source trees matching specific criteria.
You will recognise these functions because they start with the word studies_.
Now, what kind of properties can we search for in the OTOL database?
The function studies_properties() gets for us two lists, one for studies and another one for tree properties available for search.
Take a look at them:
rotl::studies_properties()
$study_properties
[1] "dc:subject" "dc:date"
[3] "ot:messages" "dc:title"
[5] "skos:changeNote" "ot:studyPublicationReference"
[7] "ot:candidateTreeForSynthesis" "ot:taxonLinkPrefixes"
[9] "treebaseId" "ot:focalCladeOTTTaxonName"
[11] "prism:modificationDate" "dc:contributor"
[13] "dc:creator" "xmlns"
[15] "ot:curatorName" "prism:number"
[17] "tb:identifier.study.tb1" "id"
[19] "ot:otusElementOrder" "ot:dataDeposit"
[21] "skos:historyNote" "ot:treesElementOrder"
[23] "prism:endingPage" "prism:section"
[25] "nexml2json" "ot:notIntendedForSynthesis"
[27] "ntrees" "treesById"
[29] "about" "prism:publicationName"
[31] "tb:identifier.study" "ot:studyYear"
[33] "otusById" "nexmljson"
[35] "ot:annotationEvents" "prism:doi"
[37] "ot:studyId" "prism:pageRange"
[39] "dc:publisher" "ot:studyPublication"
[41] "prism:volume" "tb:title.study"
[43] "ot:agents" "generator"
[45] "prism:publicationDate" "ot:tag"
[47] "ot:comment" "ot:focalClade"
[49] "prism:startingPage" "xhtml:license"
[51] "prism:creationDate" "version"
[53] "dcterms:bibliographicCitation"
$tree_properties
[1] "ot:messages" "xsi:type"
[3] "ot:nearestTaxonMRCAName" "meta"
[5] "ot:specifiedRoot" "ot:reasonsToExcludeFromSynthesis"
[7] "tb:quality.tree" "ot:branchLengthTimeUnit"
[9] "ot:nodeLabelMode" "ot:rootNodeId"
[11] "ot:inGroupClade" "ot:ottTaxonName"
[13] "ot:branchLengthDescription" "ot:studyId"
[15] "ot:MRCAName" "ot:unrootedTree"
[17] "tb:kind.tree" "tb:type.tree"
[19] "edgeBySourceId" "ot:nodeLabelDescription"
[21] "nodeById" "ot:curatedType"
[23] "ot:nearestTaxonMRCAOttId" "ot:tag"
[25] "rootedge" "label"
[27] "ntips" "tb:ntax.tree"
[29] "ot:ottId" "ot:nodeLabelTimeUnit"
[31] "ot:outGroupEdge" "ot:branchLengthMode"
[33] "ot:MRCAOttId"
As you can see, the actual values that this properties can take are not available in the output of the function. Go to the phylesystem API wiki to get them, along with an explanation of their meaning.
To get all trees with branch lengths poprotional to time we need the function
studies_find_trees(), using the property “ot:branchLengthMode” and the value “ot:time”.
It takes some time for it to get all the information, so we will not do it now.
Go to Instructor Notes later for more information on how to do this.
Search the opentree chronogram database from datelife
In the package datelife, we have implemented a workflow that extracts all studies containing information from at least two taxa.
You can get all source chronograms from an induced subtree, as long as the tip labels are in the “name” format (and not the default “name_and_id”).
datelife takes as input either a tree with tip labels as scientific names (andd not names and ids), or a vector of scientific names.
Get a Canis subtree with tip labels that do not contain the OTT id.
canis_node_subtree <- rotl::tol_subtree(node_id = canis_node_info$node_id, label = "name")
canis_node_subtree
Phylogenetic tree with 85 tips and 28 internal nodes.
Tip labels:
Canis_lupus_pallipes, Canis_lupus_chanco, Canis_lupus_baileyi, Canis_lupus_laniger, Canis_lupus_hattai, Canis_lupus_desertorum, ...
Node labels:
, , , , , , ...
Unrooted; no branch lengths.
Now, you can use that tree as input for the get_datelife_result() function.
canis_dr <- datelife::get_datelife_result(canis_node_subtree)
We have now a list of matrices storing time of lineage divergence data for all taxon pairs.
Lists are named by the study citation, so we have that information handy at all times.
Let’s explore the output.
names(canis_dr)
[1] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[2] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[3] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[4] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[5] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[6] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[7] "Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845"
canis_dr[1] # look at the first element of the list
$`Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512`
Canis rufus Canis latrans Canis simensis Canis adustus
Canis rufus 0.0 2.8 2.8 3.2
Canis latrans 2.8 0.0 2.8 3.2
Canis simensis 2.8 2.8 0.0 3.2
Canis adustus 3.2 3.2 3.2 0.0
Canis aureus 3.2 3.2 3.2 2.6
Lycalopex culpaeus 6.4 6.4 6.4 6.4
Lycalopex griseus 6.4 6.4 6.4 6.4
Lycalopex gymnocercus 6.4 6.4 6.4 6.4
Lycalopex sechurae 6.4 6.4 6.4 6.4
Lycalopex vetulus 6.4 6.4 6.4 6.4
Atelocynus microtis 6.4 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4 6.4
Chrysocyon brachyurus 6.4 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8 14.8
Canis aureus Lycalopex culpaeus Lycalopex griseus
Canis rufus 3.2 6.4 6.4
Canis latrans 3.2 6.4 6.4
Canis simensis 3.2 6.4 6.4
Canis adustus 2.6 6.4 6.4
Canis aureus 0.0 6.4 6.4
Lycalopex culpaeus 6.4 0.0 1.0
Lycalopex griseus 6.4 1.0 0.0
Lycalopex gymnocercus 6.4 1.0 1.0
Lycalopex sechurae 6.4 1.0 1.0
Lycalopex vetulus 6.4 1.4 1.4
Atelocynus microtis 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4
Chrysocyon brachyurus 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8
Lycalopex gymnocercus Lycalopex sechurae
Canis rufus 6.4 6.4
Canis latrans 6.4 6.4
Canis simensis 6.4 6.4
Canis adustus 6.4 6.4
Canis aureus 6.4 6.4
Lycalopex culpaeus 1.0 1.0
Lycalopex griseus 1.0 1.0
Lycalopex gymnocercus 0.0 1.0
Lycalopex sechurae 1.0 0.0
Lycalopex vetulus 1.4 1.4
Atelocynus microtis 6.4 6.4
Cerdocyon thous 6.4 6.4
Chrysocyon brachyurus 6.4 6.4
Lycaon pictus 6.4 6.4
Speothos venaticus 6.4 6.4
Vulpes ferrilata 14.8 14.8
Lycalopex vetulus Atelocynus microtis Cerdocyon thous
Canis rufus 6.4 6.4 6.4
Canis latrans 6.4 6.4 6.4
Canis simensis 6.4 6.4 6.4
Canis adustus 6.4 6.4 6.4
Canis aureus 6.4 6.4 6.4
Lycalopex culpaeus 1.4 6.4 6.4
Lycalopex griseus 1.4 6.4 6.4
Lycalopex gymnocercus 1.4 6.4 6.4
Lycalopex sechurae 1.4 6.4 6.4
Lycalopex vetulus 0.0 6.4 6.4
Atelocynus microtis 6.4 0.0 6.4
Cerdocyon thous 6.4 6.4 0.0
Chrysocyon brachyurus 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8
Chrysocyon brachyurus Lycaon pictus Speothos venaticus
Canis rufus 6.4 6.4 6.4
Canis latrans 6.4 6.4 6.4
Canis simensis 6.4 6.4 6.4
Canis adustus 6.4 6.4 6.4
Canis aureus 6.4 6.4 6.4
Lycalopex culpaeus 6.4 6.4 6.4
Lycalopex griseus 6.4 6.4 6.4
Lycalopex gymnocercus 6.4 6.4 6.4
Lycalopex sechurae 6.4 6.4 6.4
Lycalopex vetulus 6.4 6.4 6.4
Atelocynus microtis 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4
Chrysocyon brachyurus 0.0 6.4 6.4
Lycaon pictus 6.4 0.0 6.4
Speothos venaticus 6.4 6.4 0.0
Vulpes ferrilata 14.8 14.8 14.8
Vulpes ferrilata
Canis rufus 14.8
Canis latrans 14.8
Canis simensis 14.8
Canis adustus 14.8
Canis aureus 14.8
Lycalopex culpaeus 14.8
Lycalopex griseus 14.8
Lycalopex gymnocercus 14.8
Lycalopex sechurae 14.8
Lycalopex vetulus 14.8
Atelocynus microtis 14.8
Cerdocyon thous 14.8
Chrysocyon brachyurus 14.8
Lycaon pictus 14.8
Speothos venaticus 14.8
Vulpes ferrilata 0.0
canis_dr[length(canis_dr)] # look at the last element of the list
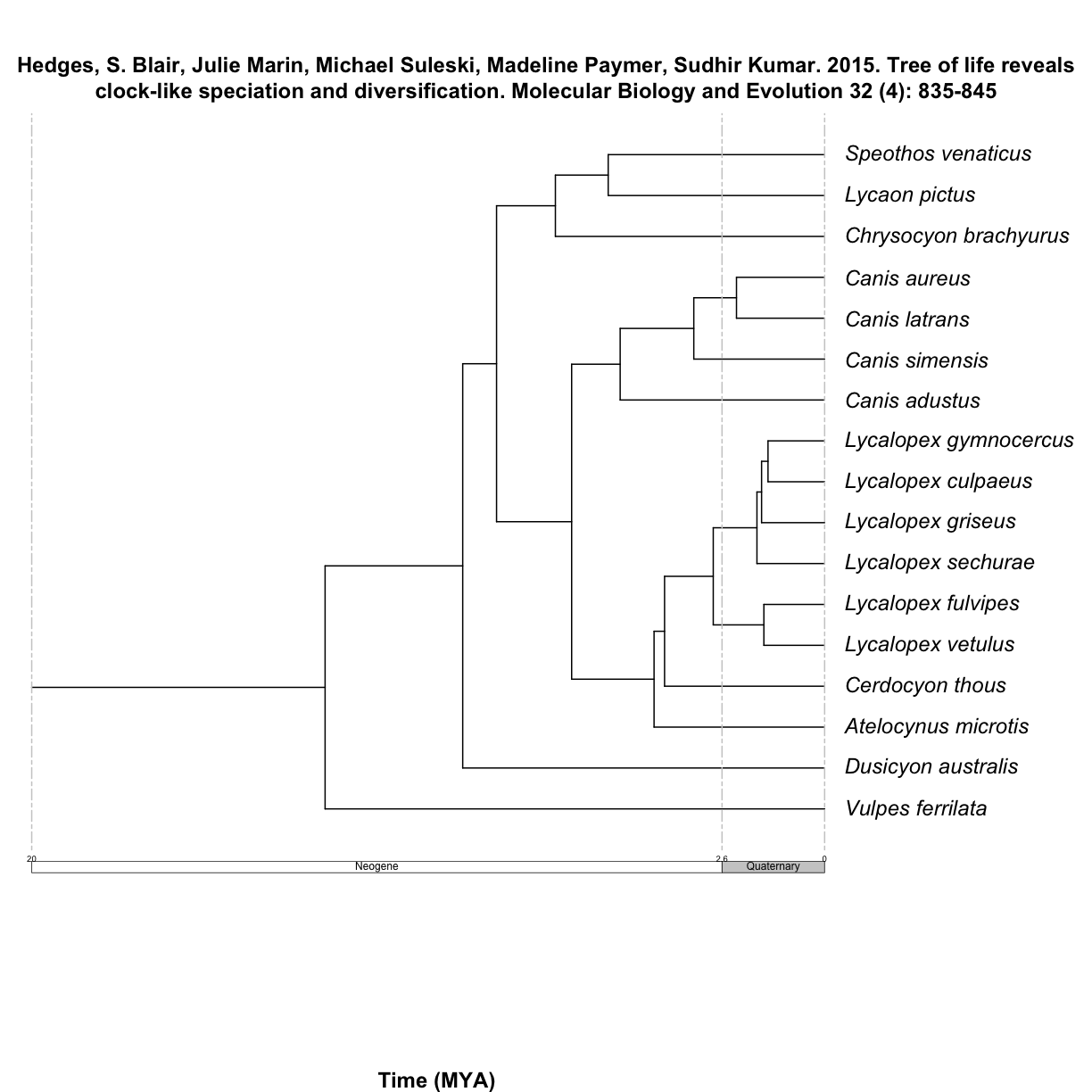
$`Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845`
Chrysocyon brachyurus Lycaon pictus Speothos venaticus
Chrysocyon brachyurus 0.00000 13.58235 13.58235
Lycaon pictus 13.58235 0.00000 10.91794
Speothos venaticus 13.58235 10.91794 0.00000
Lycalopex vetulus 16.55066 16.55066 16.55066
Lycalopex fulvipes 16.55066 16.55066 16.55066
Lycalopex culpaeus 16.55066 16.55066 16.55066
Lycalopex gymnocercus 16.55066 16.55066 16.55066
Lycalopex griseus 16.55066 16.55066 16.55066
Lycalopex sechurae 16.55066 16.55066 16.55066
Cerdocyon thous 16.55066 16.55066 16.55066
Atelocynus microtis 16.55066 16.55066 16.55066
Canis adustus 16.55066 16.55066 16.55066
Canis latrans 16.55066 16.55066 16.55066
Canis aureus 16.55066 16.55066 16.55066
Canis simensis 16.55066 16.55066 16.55066
Dusicyon australis 18.25725 18.25725 18.25725
Vulpes ferrilata 25.20000 25.20000 25.20000
Lycalopex vetulus Lycalopex fulvipes Lycalopex culpaeus
Chrysocyon brachyurus 16.550658 16.550658 16.550657
Lycaon pictus 16.550658 16.550658 16.550657
Speothos venaticus 16.550658 16.550658 16.550657
Lycalopex vetulus 0.000000 3.064400 5.618639
Lycalopex fulvipes 3.064400 0.000000 5.618639
Lycalopex culpaeus 5.618639 5.618639 0.000000
Lycalopex gymnocercus 5.618639 5.618639 2.856292
Lycalopex griseus 5.618640 5.618640 3.180365
Lycalopex sechurae 5.618640 5.618640 3.414349
Cerdocyon thous 8.074640 8.074640 8.074639
Atelocynus microtis 8.604404 8.604404 8.604403
Canis adustus 12.761959 12.761959 12.761958
Canis latrans 12.761958 12.761958 12.761957
Canis aureus 12.761958 12.761958 12.761957
Canis simensis 12.761958 12.761958 12.761957
Dusicyon australis 18.257250 18.257250 18.257249
Vulpes ferrilata 25.200000 25.200000 25.199999
Lycalopex gymnocercus Lycalopex griseus
Chrysocyon brachyurus 16.550657 16.550658
Lycaon pictus 16.550657 16.550658
Speothos venaticus 16.550657 16.550658
Lycalopex vetulus 5.618639 5.618640
Lycalopex fulvipes 5.618639 5.618640
Lycalopex culpaeus 2.856292 3.180365
Lycalopex gymnocercus 0.000000 3.180365
Lycalopex griseus 3.180365 0.000000
Lycalopex sechurae 3.414349 3.414350
Cerdocyon thous 8.074639 8.074640
Atelocynus microtis 8.604403 8.604404
Canis adustus 12.761958 12.761959
Canis latrans 12.761957 12.761958
Canis aureus 12.761957 12.761958
Canis simensis 12.761957 12.761958
Dusicyon australis 18.257249 18.257250
Vulpes ferrilata 25.199999 25.200000
Lycalopex sechurae Cerdocyon thous Atelocynus microtis
Chrysocyon brachyurus 16.550658 16.550658 16.550658
Lycaon pictus 16.550658 16.550658 16.550658
Speothos venaticus 16.550658 16.550658 16.550658
Lycalopex vetulus 5.618640 8.074640 8.604404
Lycalopex fulvipes 5.618640 8.074640 8.604404
Lycalopex culpaeus 3.414349 8.074639 8.604403
Lycalopex gymnocercus 3.414349 8.074639 8.604403
Lycalopex griseus 3.414350 8.074640 8.604404
Lycalopex sechurae 0.000000 8.074640 8.604404
Cerdocyon thous 8.074640 0.000000 8.604404
Atelocynus microtis 8.604404 8.604404 0.000000
Canis adustus 12.761959 12.761959 12.761959
Canis latrans 12.761958 12.761958 12.761958
Canis aureus 12.761958 12.761958 12.761958
Canis simensis 12.761958 12.761958 12.761958
Dusicyon australis 18.257250 18.257250 18.257250
Vulpes ferrilata 25.200000 25.200000 25.200000
Canis adustus Canis latrans Canis aureus Canis simensis
Chrysocyon brachyurus 16.55066 16.55066 16.55066 16.55066
Lycaon pictus 16.55066 16.55066 16.55066 16.55066
Speothos venaticus 16.55066 16.55066 16.55066 16.55066
Lycalopex vetulus 12.76196 12.76196 12.76196 12.76196
Lycalopex fulvipes 12.76196 12.76196 12.76196 12.76196
Lycalopex culpaeus 12.76196 12.76196 12.76196 12.76196
Lycalopex gymnocercus 12.76196 12.76196 12.76196 12.76196
Lycalopex griseus 12.76196 12.76196 12.76196 12.76196
Lycalopex sechurae 12.76196 12.76196 12.76196 12.76196
Cerdocyon thous 12.76196 12.76196 12.76196 12.76196
Atelocynus microtis 12.76196 12.76196 12.76196 12.76196
Canis adustus 0.00000 10.31455 10.31455 10.31455
Canis latrans 10.31455 0.00000 4.44640 6.60000
Canis aureus 10.31455 4.44640 0.00000 6.60000
Canis simensis 10.31455 6.60000 6.60000 0.00000
Dusicyon australis 18.25725 18.25725 18.25725 18.25725
Vulpes ferrilata 25.20000 25.20000 25.20000 25.20000
Dusicyon australis Vulpes ferrilata
Chrysocyon brachyurus 18.25725 25.2
Lycaon pictus 18.25725 25.2
Speothos venaticus 18.25725 25.2
Lycalopex vetulus 18.25725 25.2
Lycalopex fulvipes 18.25725 25.2
Lycalopex culpaeus 18.25725 25.2
Lycalopex gymnocercus 18.25725 25.2
Lycalopex griseus 18.25725 25.2
Lycalopex sechurae 18.25725 25.2
Cerdocyon thous 18.25725 25.2
Atelocynus microtis 18.25725 25.2
Canis adustus 18.25725 25.2
Canis latrans 18.25725 25.2
Canis aureus 18.25725 25.2
Canis simensis 18.25725 25.2
Dusicyon australis 0.00000 25.2
Vulpes ferrilata 25.20000 0.0
Get your chronograms
Then, it is really easy to go from a matrix to a tree, using the function summarize_datelife_result() with the option summary_format = "phylo_all". Note the printed output returns a summary of taxa that have branch length information in the database.
canis_phylo_all <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_all")
Source chronograms from:
1: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
2: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
3: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
4: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
5: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
6: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
7: Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845
Input taxa presence across source chronograms:
taxon chronograms
1 Canis rufus 3/7
2 Canis latrans 7/7
3 Canis simensis 7/7
4 Canis adustus 7/7
5 Canis aureus 7/7
6 Lycalopex culpaeus 7/7
7 Lycalopex griseus 7/7
8 Lycalopex gymnocercus 7/7
9 Lycalopex sechurae 7/7
10 Lycalopex vetulus 7/7
11 Atelocynus microtis 7/7
12 Cerdocyon thous 7/7
13 Chrysocyon brachyurus 7/7
14 Lycaon pictus 7/7
15 Speothos venaticus 7/7
16 Vulpes ferrilata 7/7
17 Dusicyon australis 4/7
18 Lycalopex fulvipes 4/7
Input taxa completely absent from source chronograms:
taxon
1 Canis himalayensis
2 Canis indica
3 Canis environmental sample
4 Canis anthus
5 Canis antarticus
6 Canis dingo
7 Canis ameghinoi
8 Canis argentinus
9 Canis cautleyi
10 Canis chanco
11 Canis chrysurus
12 Canis curvipalatus
13 Canis dukhunensis
14 Canis etruscus
15 Canis gezi
16 Canis himalaicus
17 Canis kokree
18 Canis lanka
19 Canis lateralis
20 Canis naria
21 Canis nehringi
22 Canis pallipes
23 Canis palustris
24 Canis peruanus
25 Canis primaevus
26 Canis sladeni
27 Canis tarijensis
28 Canis ursinus
29 Canis ferox
30 Canis lupus pallipes
31 Canis lupus chanco
32 Canis lupus baileyi
33 Canis lupus laniger
34 Canis lupus hattai
35 Canis lupus desertorum
36 Canis lupus familiaris
37 Canis lupus mogollonensis
38 Canis lupus hodophilax
39 Canis lupus dingo
40 Canis lupus labradorius
41 Canis lupus signatus
42 Canis lupus lupus
43 Canis lupus lycaon
44 Canis lupus lupaster
45 Canis lupus campestris
46 Canis lupus arctos
47 Canis lupus variabilis
48 Canis lupus orion
49 Canis dirus
50 Canis armbrusteri
51 Speothos pacivorus
52 Cuon primaevus
53 Cuon javanicus
54 Cuon stehlini
55 Cuon alpinus lepturus
56 Canis edwardii
57 Canis lepophagus
58 Canis cedazoensis
59 Canis davisi
60 Dusicyon avus
61 Dusicyon darwini
62 Dusicyon gymnocercus
63 Dusicyon proplatensis
64 Lycalopex sp. Fuegian dog
65 Lycalopex fulvicaudus
66 Canis mesomelas elongae
67 Cerdocyon ensenadensis
Plot your results
To plot the resulting tree, you can use the plot.phylo() function from ape.
You can also use the function plot_phylo_all(), that adds the study citation as title and a geostratigraphic axis.
datelife::plot_phylo_all(trees = canis_phylo_all)






Key Points
datelife stores all chronograms from the Open Tree of Life phylesystem.
chronograms are stored in the
opentree_chronogramsobject.source chronograms are retrieved at the species level only (for now).