1. Finding your taxa in the Open Tree of Life Taxonomy
Overview
Teaching: 5 min
Exercises: 5 minQuestions
What is the Open Tree Taxonomy?
What are OTT ids?
What does TNRS stand for?
Objectives
Getting OTT ids for some taxa.
Understanding TNRS, approximate matching and its sensitivity.
The Open Tree Taxonomy (OTT from now on) synthesizes taxonomic information from different sources and assigns each taxon a unique identifier, which we refer to as the OTT id. To interact with the OTT (and other Open Tree of Life services) using R, we will learn how to use the functions from the rotl package. If you don’t know if you have the package installed, go to setup and follow the instructions there.
To deal with synonyms and scientific name misspellings, the Open Tree Taxonomy uses
the Taxonomic Name Resolution Service (TNRS
from now on), that allows linking scientific names to a unique OTT id, while dealing
with misspellings, synonyms and scientific name variants. The functions from rotl that interact
with the OTT start with “tnrs_”.
Note: Going from a common name to a scientific name
TNRS only deals with scientific names. If you want to work with common names, you will have to use a service that can get the scientific name of a list of common names. There are no functions in
rotlto deal with this. We know of at least two places that have implemented this otherwise. The phylotastic project has implemented a common name to scientific name service that is also available in the r package rphylotastic. The OneZoom project has also developed a service that matches common names from the Encyclopedia of Life to scientific names.
To get OTT ids for a set of taxa we will use the function tnrs_match_names().
This function takes a character vector of one or more taxon scientific names as main argument.
Hands on! Run TNRS
Do a
tnrs_match_names()run on the amphibians, the genus of the dog (Canis), the genus of the cat (Felis), the family of dolphins (Delphinidae) and the class of birds (Aves). Save the output to an object namedresolved_names.my_taxa <- c("amphibians", "canis", "felis", "delphinidae", "ave") resolved_names <- rotl::tnrs_match_names(names = my_taxa)You can try different misspellings and synonyms of your taxa to see TNRS in action.
Ok, we ran the function successfully. Now Let’s explore its output.
resolved_names
search_string unique_name approximate_match ott_id is_synonym flags number_matches
1 amphibians Amphibia TRUE 544595 FALSE 6
2 canis Canis FALSE 372706 FALSE 2
3 felis Felis FALSE 563165 FALSE 1
4 delphinidae Delphinidae FALSE 698406 FALSE 1
5 ave Aves TRUE 81461 FALSE 6
The output of the function is a data table. In R (and other coding languages), objects are assigned to classes to make their manipulation with other functions much easier.
A class is basically a data structure that is the same among all objects that belong to the same class.
Let’s explore the class of the tnrs_match_names() output.
class(resolved_names)
[1] "match_names" "data.frame"
As you can see, an object can belong to one or more classes.
Indeed, R is telling us that the output of tnrs_match_names() is a data frame (a type of table) and a ‘match_names’ object, which is in turn a data frame with exactly 7 named columns: search_string, unique_name, approximate_match, ott_id, is_synonym, flags, and number_matches.
Ok, so we know now what is the data structure of the ‘match_names’ object, but what kind of data can I find in this class of objects?
Kinds of data stored in a ‘match_names’ object
You should have a good idea by now of what type of data is stored in the ott_ids column.
Can you guess what type of data is displayed in the column search_string and unique_name?
How about is_synonym?
The column approximate_match tells us whether the unique name was inferred from the search string using approximate matching (TRUE) or not (FALSE).
Finally, the flags column tells us if our unique name has been flagged in the OTT
(TRUE) or not (FALSE), and the type of flag if any. Flags are markers that indicate if the taxon in question should be included in further analyses of the Open Tree workflow. You can read more about flags in the wiki.
Pro tip 1.1: Looking at “hidden” elements of a data object
The ‘match_names’ object actually has more data that is not exposed on the screen and is not part of the main data structure. This “hidden” data is stored in the attributes of the object. All objects have at least one attribute, the class. Attributes can be accesed with the function
attributes(), and are stored as a named list.Explore the attributes of the ‘match_names’ object
names(attributes(resolved_names))[1] "names" "row.names" "original_order" [4] "original_response" "match_id" "has_original_match" [7] "class"Look at the attributes of other objects:
attributes(my_taxa)NULLAs you can see there are many more attributes in a ‘match_names’ object than in simpler objects.
Now we know what kind of data is retrieved by the tnrs_match_names() function. Pretty cool!
Finally, how can I extract specific pieces of data from my object to use elsewhere?
Extracting data from a ‘match_names’ object
It is easy to access elements from a ‘match_names’ object using regular indexing. For example, using the column number, we can extract all elements from a certain column. Let’s extract all data from the second column:
resolved_names[,2]
[1] "Amphibia" "Canis" "Felis" "Delphinidae" "Aves"
We can also use the name of the column so we do not have to remember its position:
resolved_names[,"unique_name"]
[1] "Amphibia" "Canis" "Felis" "Delphinidae" "Aves"
Because it is a ‘data.frame’, we can also access the values of any column by using the “$” and the column name to index it, like this:
resolved_names$unique_name
[1] "Amphibia" "Canis" "Felis" "Delphinidae" "Aves"
The ‘match_names’ object has a relatively simple structure that is easy to explore and mine.
We will see later that the outputs of other rotl functions are way more complicated
and accesing their elements requires a lot of hacking. Fortunately, the rotl creators have
added some functions that allow interacting with these complicated outputs.
The functions unique_name(), ott_id(), and flags() extract values from the
respective columns of a ‘match_names’ object, in the form of a list instead of a vector.
To extract data from the other columns there are no specialized functions, so you will have to index.
Hands on! Extract the OTT ids from a ‘match_names’ object
You now have a ‘match_names’ object that we called
resolved_names. There are at least two ways to extract the OTT ids from it. Can you figure them out? Store them in an object we will callmy_ott_ids.Hint: You can find one solution by browsing the rotl package documentation to find a function that will do this for a ‘match_names’ object.
You will find a second solution by using your knowledge on data frames and tables to extract the data from the
ott_idcolumn.Look at some solutions
Get the OTT ids as a list, with the function
ott_id():my_ott_ids <- rotl::ott_id(resolved_names) # rotl:::ott_id.match_names(resolved_names) is the same. my_ott_ids$Amphibia [1] 544595 $Canis [1] 372706 $Felis [1] 563165 $Delphinidae [1] 698406 $Aves [1] 81461 attr(,"class") [1] "otl_ott_id" "list"Or, get the OTT ids as a vector:
my_ott_ids <- resolved_names$ott_id # or resolved_names[, "ott_id"] my_ott_ids[1] 544595 372706 563165 698406 81461
There are no specialized functions to extract values from a row of a ‘match_names’ object, so we have to do some indexing. You can get values from all columns of one row:
resolved_names[1,]
search_string unique_name approximate_match ott_id is_synonym flags
1 amphibians Amphibia TRUE 544595 FALSE
number_matches
1 6
Or get just one specific value from a certain column, using the column name:
resolved_names[1,"unique_name"]
[1] "Amphibia"
Or using the column position:
resolved_names[1,2]
[1] "Amphibia"
Hack: Name the rows of your ‘match_names’ object
To facilitate the use of OTT ids later, you can name the rows of your ‘match_names’ object using the function
rownames().You can name them whatever you want. For example, you can use the
unique_nameidentifier:rownames(resolved_names) <- resolved_names$unique_nameOr simply call them something short that makes sense to you and is easy to remember:
rownames(resolved_names) <- c("amphs", "dogs", "cats", "flippers", "birds")This will facilitate accessing elements of the ‘match_names’ object by allowing to just use the row name as row index (instead of a number).
There are at least two ways to do this.
You can use the “$” to acces a named column of the data frame:
resolved_names["flippers",]$ott_id[1] 698406Or you can use the column name as column index:
resolved_names["flippers","ott_id"][1] 698406In both cases, you will get the OTT id of the Delphinidae. Cool!
Pro tip 1.2: Extract data from the attributes of a ‘match_names’ object
On the previous
pro tipwe saw that there is more data stored in the attributes of the ‘match_names’ object. The structure of this data is complicated and extracting it requires some hacking. There is one inbuilt function in the packagerotlthat will extract the synonyms from the attributes of a ‘match_names’ object.The function
synonyms()rotl::synonyms(resolved_names)$Amphibia [1] "Lissamphibia" "Amphibia" $Canis [1] "Vulpicanis" "Lupulella" "Chaon" "Dasycyon" "Simenia" [6] "Lupulus" "Dimenia" "Alopedon" "Thos" "Schaeffia" [11] "Canix" "Jacalius" "Mamcanisus" "Sacalius" "Oxygous" [16] "Neocyon" "Lupus" "Aenocyon" "Canis" "Alopsis" [21] "Oxygonus" "Lyciscus" "Oreocyon" "Dieba" $Felis [1] "Felis" "Felix" "Microfelis" "Trichaelurus" "Poliailurus" [6] "Chaus" "Catolynx" "Felia" "Folis" "Otocolobus" [11] "Otocalobus" "Mamfelisus" "Otailurus" "Eremaelurus" "Avitofelis" [16] "Octolobus" "Ictalurus" "Catus" "Octalobus" $Delphinidae [1] "Orcinae" "Orcini" "Orcadae" [4] "Orcaelidae" "Trispondylus kleini" "Stenidae" [7] "Globicephalidae" "Orcininae" "Delphinusideae" [10] "Globidelphinidae" "Delphinidae" "Orcaellidae" [13] "Orcadina" "Delphinapteridae" "Delphinoidae" [16] "Grampidelphidae" "Trispondylus" "Cephalorhynchinae" [19] "Tursiops miocaenus" "Grampidae" "Globiocephalidae" [22] "Steno cudmorei" $Aves [1] "Aves" "avian" "Lophorus" "Lepturus" "Phyllomanes" attr(,"class") [1] "otl_synonyms" "list"Neat!
There you go! Now we know how to get OTT ids from a bunch of taxa of interest. Let’s see what we can do with these on the next section.
Key Points
OTT ids are the Open Tree of Life Taxonomy handlers; they identify taxa.
You can go from a scientific name to an OTT id with TNRS matching.
You cannot go from a common name to OTT id with the Open Tree of Life tools.
2. Getting a piece of the Synthetic Open Tree of Life
Overview
Teaching: 5 min
Exercises: 5 minQuestions
What is the synthetic Open Tree of Life?
How do I interact with it?
Why is my taxon not in the tree?
Objectives
Get an induced subtree
Get a subtree
The synthetic Open Tree of Life (synthetic OTOL from now on) summarizes information from 1216 trees from 1162 peer-reviewed and published studies, that have been uploaded to the OTOL database through a curator system.
Functions from the rotl package that interact with the synthetic OTOL start with “tol_”.
To access general information about the current synthetic OTOL, we can use the function tol_about(). This function requires no argument.
rotl::tol_about()
OpenTree Synthetic Tree of Life.
Tree version: opentree12.3
Taxonomy version: 3.2draft9
Constructed on: 2019-12-23 11:41:23
Number of terminal taxa: 2391916
Number of source trees: 1216
Number of source studies: 1162
Source list present: false
Root taxon: cellular organisms
Root ott_id: 93302
Root node_id: ott93302
This is nice!
As you can note, the current synthetic OTOL was created not too long ago, on 2019-12-23 11:41:23.
This is also telling us that there are currently more than 2 million tips on the synthetic OTOL.
It is indeed a large tree. So, what if we just want a small piece of the whole synthetic OTOL?
Well, now that we have some interesting taxon OTT ids, we can easily do this.
Getting an induced subtree
The function tol_induced_subtree() allows us to get a tree of taxa from different taxonomic ranks.
my_tree <- rotl::tol_induced_subtree(resolved_names$ott_id)
Warning in collapse_singles(tr, show_progress): Dropping singleton nodes
with labels: Mammalia ott244265, Theria (subclass in Deuterostomia)
ott229558, Eutheria (in Deuterostomia) ott683263, Boreoeutheria ott5334778,
Laurasiatheria ott392223, mrcaott1548ott6790, mrcaott1548ott3607484,
mrcaott1548ott4942380, mrcaott1548ott4942547, mrcaott1548ott3021, Artiodactyla
ott622916, mrcaott1548ott21987, mrcaott1548ott5256, mrcaott5256ott4944931,
Whippomorpha ott7655791, Cetacea ott698424, mrcaott5256ott3615450,
mrcaott5256ott44568, Odontoceti ott698417, mrcaott5256ott5269,
mrcaott5269ott6470, mrcaott5269ott47843, mrcaott47843ott194312,
mrcaott4697ott263949, Carnivora ott44565, Caniformia ott827263,
Canidae ott770319, mrcaott47497ott3612617, mrcaott47497ott3612529,
mrcaott47497ott3612596, mrcaott47497ott3612516, mrcaott47497ott3612589,
mrcaott47497ott3612591, mrcaott47497ott3612592, mrcaott47497ott77889,
Feliformia ott827259, mrcaott6940ott19397, mrcaott19397ott194349, Felidae
ott563159, mrcaott54737ott660452, mrcaott54737ott86170, mrcaott54737ott86175,
mrcaott54737ott442049, mrcaott54737ott86162, mrcaott54737ott86166, Sauropsida
ott639642, Sauria ott329823, mrcaott246ott4128455, mrcaott246ott4127082,
mrcaott246ott4129629, mrcaott246ott4142716, mrcaott246ott4126667,
mrcaott246ott1662, mrcaott246ott2982, mrcaott246ott31216, mrcaott246ott4947920,
mrcaott246ott4127428, mrcaott246ott4126230, mrcaott246ott4127421,
mrcaott246ott664349, mrcaott246ott4126505, mrcaott246ott4127015,
mrcaott246ott4129653, mrcaott246ott4127541, mrcaott246ott4946623,
mrcaott246ott4126482, mrcaott246ott4128105, mrcaott246ott4127288,
mrcaott246ott4132146, mrcaott246ott3602822, mrcaott246ott4143599,
mrcaott246ott3600976, mrcaott246ott4132107, Aves ott81461, Neognathae
ott241846, mrcaott246ott5481, mrcaott246ott5021, mrcaott246ott7145,
mrcaott246ott5272, mrcaott5272ott9830, mrcaott9830ott86672, mrcaott9830ott90560,
mrcaott9830ott18206, mrcaott18206ott60413, Sphenisciformes ott494366
Note: What does this warning mean?
This warning has to do with the way the synthetic OTOL is generated. You can look at the overview of the synthesis algorithm for more information.
Let’s look at the output of tol_induced_subtree().
my_tree
Phylogenetic tree with 5 tips and 4 internal nodes.
Tip labels:
[1] "Delphinidae_ott698406" "mrcaott47497ott110766" "Felis_ott563165"
[4] "Spheniscidae_ott494367" "Amphibia_ott544595"
Node labels:
[1] "Tetrapoda ott229562" "Amniota ott229560" "mrcaott1548ott4697"
[4] "mrcaott4697ott6940"
Rooted; no branch lengths.
R is telling us that we have a rooted tree with no branch lengths and 5 tips. If we check the class of the output, we will verify that it is a ‘phylo’ object.
class(my_tree)
[1] "phylo"
A ‘phylo’ object is a data structure that stores the necessary information to build a tree.
There are several functions from different packages to plot trees or ‘phylo’ objects in R (e.g., phytools). For now, we will use the one from the legendary ape package plot.phylo():
ape::plot.phylo(my_tree, cex = 2) # or just plot(my_tree, cex = 2)
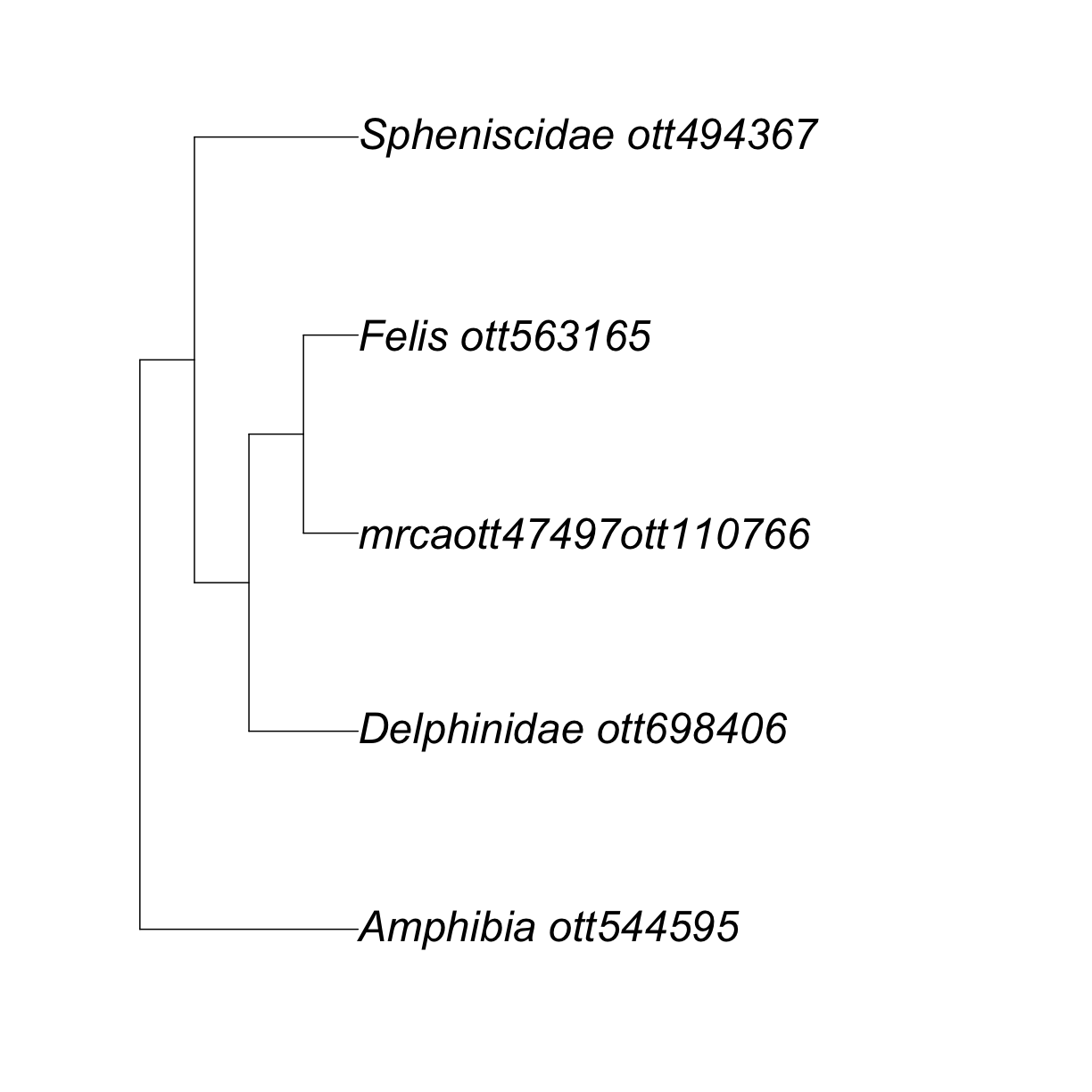
This is cool!
But, why oh why did my Canis disappear? 😢
Well, it did not actually disappear, it was replaced by the label “mrcaott47497ott110766”.
We will explain why this happens in the next section.
Now, what if you want a piece of the synthetic OTOL containing all descendants of your taxa of interest?
Getting a subtree of one taxon
We can extract a subtree of all descendants of one taxon at a time using the function tol_subtree() and an OTT id of your choosing.
Let’s extract a subtree of all amphibians.
First, get its OTT id. It is already stored in our resolved_names object:
amphibia_ott_id <- resolved_names["Amphibia",]$ott_id
Or, you can run the function tnrs_match_names() again if you want.
amphibia_ott_id <- rotl::tnrs_match_names("amphibians")$ott_id
Now, extract the subtree from the synthetic OTOL using tol_subtree().
amphibia_subtree <- rotl::tol_subtree(ott_id = resolved_names["Amphibia",]$ott_id)
Let’s look at the output:
amphibia_subtree
Phylogenetic tree with 10012 tips and 3100 internal nodes.
Tip labels:
Odorrana_geminata_ott114, Odorrana_supranarina_ott14375, Odorrana_narina_ott14379, Odorrana_amamiensis_ott14384, Odorrana_utsunomiyaorum_ott14377, Odorrana_swinhoana_ott14392, ...
Node labels:
Amphibia ott544595, Batrachia ott471197, Anura ott991547, , , , ...
Unrooted; no branch lengths.
This is a large tree! We will have a hard time plotting it.
Now, let’s extract a subtree for the genus Canis. It should be way smaller!
subtree <- rotl::tol_subtree(resolved_names["Canis",]$ott_id)
Error: HTTP failure: 400
list(contesting_trees = list(`ot_278@tree1` = list(attachment_points = list(list(children_from_taxon = list("node242"), parent = "node241"), list(children_from_taxon = list("node244"), parent = "node243"), list(children_from_taxon = list("node262"), parent = "node255"), list(children_from_taxon = list("node270"), parent = "node267"))), `ot_328@tree1` = list(attachment_points = list(list(children_from_taxon = list("node519"), parent = "node518"), list(children_from_taxon = list("node523"), parent = "node522")))),
mrca = "mrcaott47497ott110766")[/v3/tree_of_life/subtree] Error: node_id was not found (broken taxon).
😱 😱 😱
What does this error mean??
A “broken” taxon error usually happens when phylogenetic information does not match taxonomic information.
For example, extinct lineages are sometimes phylogenetically included within a taxon but are taxonomically excluded, making the taxon appear as paraphyletic.
On the Open Tree of Life browser, we can still get to the subtree (check it out here).
From R, we will need to do something else first. We will get to that on the next episode.
Key Points
OTT ids and node ids allow us to interact with the synthetic OTOL.
Portions of the synthetic OTOL can be extracted from a single OTT id or from a bunch of them.
It is not possible to get a subtree from an OTT id that is not in the synthetic tree.
3. Dealing with "broken" and "invalid" taxa
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I detect a broken taxon?
Objectives
Use the function is_in_tree()
Understand outputs from those functions
We say that a taxon is “broken” when its OTT id is not assigned to a node in the synthetic tree. As mentioned before, this happens when the OTT id belongs to a taxon that is not monophyletic in the synthetic tree. This is the reason why we get an error when we try to get a synthetic subtree including that OTT id: it is not in the tree.
There is a way to find out that a group is “broken” before trying to get the subtree and getting an error.
rotl::is_in_tree(resolved_names["Canis",]$ott_id)
[1] FALSE
Indeed, our Canis is not in the synthetic OTOL. To extract a subtree of a “broken” taxon, we have some options. But we will focus on one.
Getting the MRCA of a taxon
The function tol_node_info() gets for you all relevant information of the node that is the ancestor or MRCA of a taxon. That also includes the actual node id.
canis_node_info <- rotl::tol_node_info(resolved_names["Canis",]$ott_id)
canis_node_info
OpenTree node.
Node id: mrcaott47497ott110766
Number of terminal descendants: 85
Is taxon: FALSE
Let’s explore the class of the output.
class(canis_node_info)
[1] "tol_node" "list"
So we have an object of class ‘list’ and ‘tol_node’. When we printed it, we got some information. But we do not know how much information might not be “printed” to screen.
Let’s use the functions str() or ls() to check out the data strcture of our ‘tol_node’ object.
str(canis_node_info)
List of 8
$ node_id : chr "mrcaott47497ott110766"
$ num_tips : int 85
$ query : chr "ott372706"
$ resolves :List of 1
..$ pg_2812@tree6545: chr "node1135827"
$ source_id_map:List of 5
..$ ot_278@tree1 :List of 3
.. ..$ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960"
.. ..$ study_id: chr "ot_278"
.. ..$ tree_id : chr "tree1"
..$ ot_328@tree1 :List of 3
.. ..$ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960"
.. ..$ study_id: chr "ot_328"
.. ..$ tree_id : chr "tree1"
..$ pg_1428@tree2855:List of 3
.. ..$ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960"
.. ..$ study_id: chr "pg_1428"
.. ..$ tree_id : chr "tree2855"
..$ pg_2647@tree6169:List of 3
.. ..$ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960"
.. ..$ study_id: chr "pg_2647"
.. ..$ tree_id : chr "tree6169"
..$ pg_2812@tree6545:List of 3
.. ..$ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960"
.. ..$ study_id: chr "pg_2812"
.. ..$ tree_id : chr "tree6545"
$ supported_by :List of 2
..$ ot_278@tree1: chr "node233"
..$ ot_328@tree1: chr "node495"
$ synth_id : chr "opentree12.3"
$ terminal :List of 2
..$ pg_1428@tree2855: chr "node610132"
..$ pg_2647@tree6169: chr "ott247333"
- attr(*, "class")= chr [1:2] "tol_node" "list"
This is telling us that tol_node_info() extracted 8 different pieces of information from my node.
Right now we are only interested in the node ir. Where do you think it is?
Hands on! Get the node id of Canis MRCA
Extract it from your
canis_node_infoobject and call itcanis_node_id.canis_node_id <- canis_node_info$node_id
Pro tip 3.1: Get the node id of the MRCA of a group of OTT ids
Sometimes you want the MRCA of a bunch of lineages. The function
tol_mrca()gets the node if of the MRCA of a group of OTT ids.Can you use it to get the mrca of Canis?
The node that contains Canis is “mrcaott47497ott110766”.
Getting a subtree using a node id instead of the taxon OTT id
Now that we have a node id, we can use it to get a subtree with tol_subtree(), using the argument node_id.
canis_node_subtree <- rotl::tol_subtree(node_id = canis_node_id)
ape::plot.phylo(canis_node_subtree, cex = 1.2)
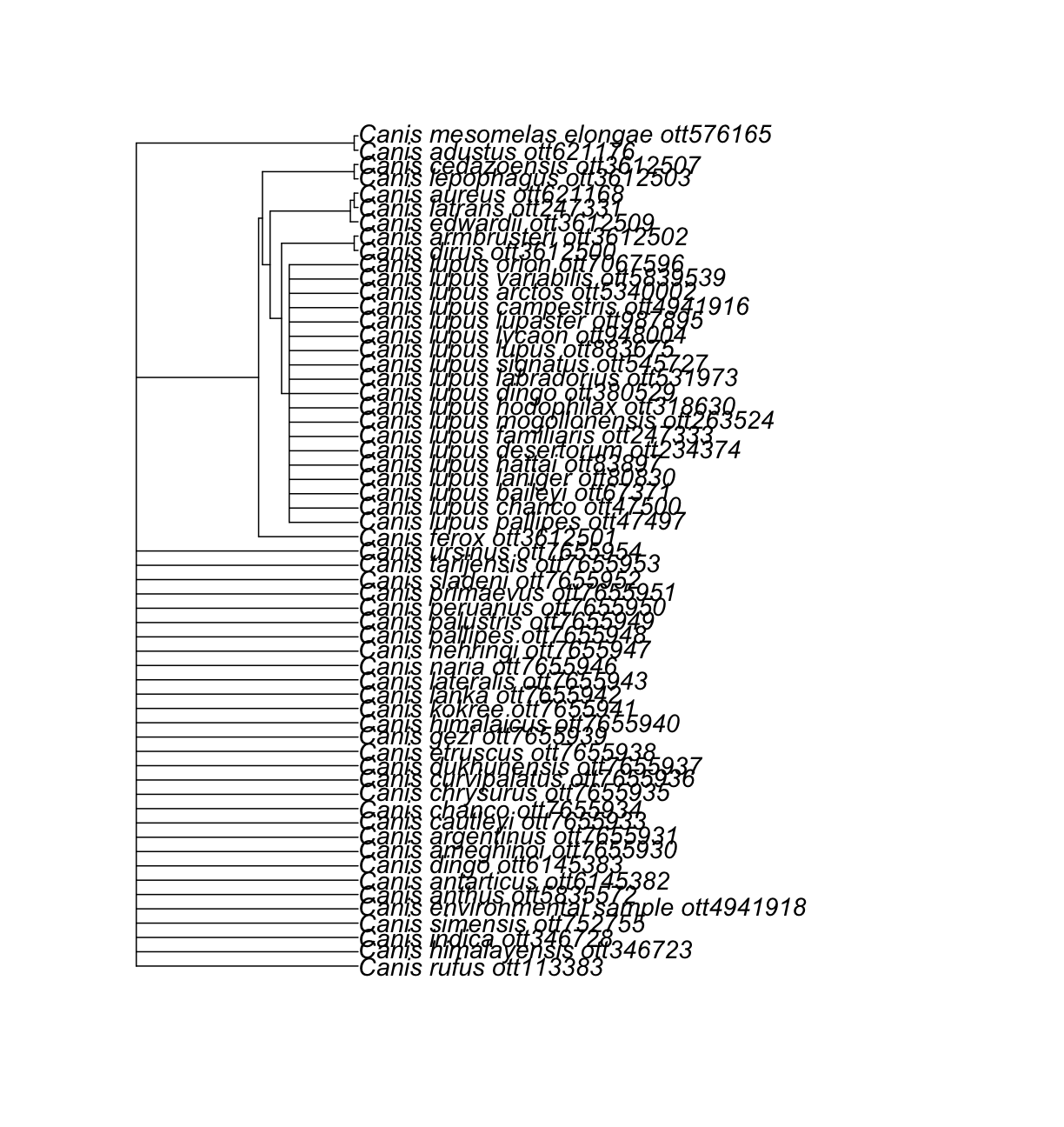
Nice! We got a subtree of 85 tips, containing all descendants from the node that also contains Canis.
This includes species assigned to genera other than Canis.
Note: Get an induced subtree of taxonomic children
It might seem non phylogenetic, but what if I really, really need a tree containing species within the genus Canis only?
We can get the OTT ids of the taxonomic children of our taxon of interest and use the function
tol_induced_subtree().So, here is my hack, enjoy!
First, get the taxonomic children.
canis_taxonomy <- rotl::taxonomy_subtree(resolved_names["Canis",]$ott_id)canis_taxonomy$tip_label [1] "Canis_dirus_ott3612500" [2] "Canis_anthus_ott5835572" [3] "Canis_rufus_ott113383" [4] "Canis_simensis_ott752755" [5] "Canis_aureus_ott621168" [6] "Canis_mesomelas_elongae_ott576165" [7] "Canis_adustus_ott621176" [8] "unclassified_Canis_ott7655955" [9] "Canis_latrans_ott247331" [10] "Canis_lupus_baileyi_ott67371" [11] "Canis_lupus_laniger_ott80830" [12] "Canis_lupus_orion_ott7067596" [13] "Canis_lupus_hodophilax_ott318630" [14] "Canis_lupus_signatus_ott545727" [15] "Canis_lupus_arctos_ott5340002" [16] "Canis_lupus_mogollonensis_ott263524" [17] "Canis_lupus_variabilis_ott5839539" [18] "Canis_lupus_lupus_ott883675" [19] "Canis_lupus_campestris_ott4941916" [20] "Canis_lupus_lycaon_ott948004" [21] "Canis_lupus_pallipes_ott47497" [22] "Canis_lupus_chanco_ott47500" [23] "Canis_lupus_x_Canis_lupus_familiaris_ott4941915" [24] "Canis_lupus_desertorum_ott234374" [25] "Canis_lupus_familiaris_ott247333" [26] "Canis_lupus_dingo_ott380529" [27] "Canis_lupus_labradorius_ott531973" [28] "Canis_lupus_hattai_ott83897" [29] "Canis_lupus_lupaster_ott987895" [30] "Canis_himalayensis_ott346723" [31] "Canis_indica_ott346728" [32] "Canis_environmental_samples_ott4941917" [33] "Canissp.KEB-2016ott5925604" [34] "Canis_sp._CANInt1_ott470950" [35] "'Canissp.Russia/33" [36] "500ott5338950'" [37] "Canis_sp._ott247325" [38] "'Canissp.Belgium/36" [39] "000ott5338951'" [40] "Canis_environmental_sample_ott4941918" [41] "Canis_morenis_ott6145387" [42] "Canis_niger_ott6145388" [43] "Canis_palaeoplatensis_ott6145390" [44] "Canis_osorum_ott6145389" [45] "Canis_thooides_ott6145392" [46] "Canis_antarcticus_ott6145381" [47] "Canis_proplatensis_ott6145391" [48] "Canis_feneus_ott6145384" [49] "Canis_geismarianus_ott6145385" [50] "Canis_ameghinoi_ott7655930" [51] "Canis_nehringi_ott7655947" [52] "Canis_palustris_ott7655949" [53] "Canis_lanka_ott7655942" [54] "Canis_pallipes_ott7655948" [55] "Canis_gezi_ott7655939" [56] "Canis_montanus_ott7655945" [57] "Canis_primaevus_ott7655951" [58] "Canis_chrysurus_ott7655935" [59] "Canis_dukhunensis_ott7655937" [60] "Canis_kokree_ott7655941" [61] "Canis_sladeni_ott7655952" [62] "Canis_himalaicus_ott7655940" [63] "Canis_chanco_ott7655934" [64] "Canis_curvipalatus_ott7655936" [65] "Canis_lateralis_ott7655943" [66] "Canis_argentinus_ott7655931" [67] "Canis_tarijensis_ott7655953" [68] "Canis_naria_ott7655946" [69] "Canis_peruanus_ott7655950" [70] "Canis_cautleyi_ott7655933" [71] "Canis_ursinus_ott7655954" [72] "Canis_armbrusteri_ott3612502" [73] "Canis_ferox_ott3612501" [74] "Canis_lepophagus_ott3612503" [75] "Canis_edwardii_ott3612509" [76] "Canis_apolloniensis_ott3612508" [77] "Canis_cedazoensis_ott3612507" [78] "Canis_primigenius_ott3612506" [79] "Canis_lydekkeri_ott7655944" [80] "Canis_arnensis_ott7655932" [81] "Canis_antarticus_ott6145382" [82] "Canis_dingo_ott6145383" [83] "Canis_etruscus_ott7655938" [84] "Canis_spelaeus_ott3612504" $edge_label [1] "Canis_mesomelas_ott666235" "Canis_lupus_ott247341" [3] "Canis_ott372706"Now, extract the OTT ids.
canis_taxonomy_ott_ids <- datelife::extract_ott_ids(x = canis_taxonomy$tip_label)After extracting ott ids, there are some non numeric elements:Canissp.KEB-2016ott5925604 'Canissp.Russia/33 500ott5338950' 'Canissp.Belgium/36 000ott5338951'NAs removed.Try to get an induced subtree of Canis taxonomic children.
canis_taxonomy_subtree <- rotl::tol_induced_subtree(canis_taxonomy_ott_ids)Error: HTTP failure: 400 [/v3/tree_of_life/induced_subtree] Error: node_id 'ott3612504' was not found!list(ott247325 = "pruned_ott_id", ott3612504 = "pruned_ott_id", ott3612506 = "pruned_ott_id", ott3612508 = "pruned_ott_id", ott470950 = "pruned_ott_id", ott4941915 = "pruned_ott_id", ott4941917 = "pruned_ott_id", ott6145381 = "pruned_ott_id", ott6145384 = "pruned_ott_id", ott6145385 = "pruned_ott_id", ott6145387 = "pruned_ott_id", ott6145388 = "pruned_ott_id", ott6145389 = "pruned_ott_id", ott6145390 = "pruned_ott_id", ott6145391 = "pruned_ott_id", ott6145392 = "pruned_ott_id", ott7655932 = "pruned_ott_id", ott7655944 = "pruned_ott_id", ott7655945 = "pruned_ott_id", ott7655955 = "pruned_ott_id")It is often not possible to get an induced subtree of all taxonomic children from a taxon, because some of them will not make it to the synthetic tree.
To verify which ones are giving us trouble, we can use the function
is_in_tree()again.canis_in_tree <- sapply(canis_taxonomy_ott_ids, rotl::is_in_tree) # logical vector canis_taxonomy_ott_ids_intree <- canis_taxonomy_ott_ids[canis_in_tree] # extract ott ids in treeNow get the tree.
canis_taxonomy_subtree <- rotl::tol_induced_subtree(canis_taxonomy_ott_ids_intree)Plot it.
ape::plot.phylo(canis_taxonomy_subtree, cex = 1.2)
There! We have a synthetic subtree (derived from phylogenetic information) containing only the taxonomic children of Canis.
What if I want a subtree of certain taxonomic ranks withing my group? Go to the next episode and find out how you can do this!
Key Points
It is not possible to get a subtre from an OTT id that is not in the synthetic tree.
OTT ids and node ids allow us to interact with the synthetic OTOL.
4. Getting an induced subtree of all taxa within a taxonomic rank
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I get all taxa from a certain rank?
Objectives
Get an induced subtree from all taxa of a certain taxonomic rank.
There is not a specific function to get all taxa from a given rank.
Of course, we can always hack our way through it.
In the package datelife, we have implemented a function called get_ott_children(), that extracts OTT ids of all taxa from a rank specified by the argument ott_rank.
Let’s get all amphibian families.
amphibia_families <- datelife::get_ott_children(ott_ids = resolved_names["Amphibia",]$ott_id, ott_rank = "family")
Now, get the induced subtree using the amphibian families’ OTT ids.
amphibia_families_subtree <- rotl::tol_induced_subtree(amphibia_families$Amphibia$ott_id)
Look at the output.
amphibia_families_subtree
Phylogenetic tree with 61 tips and 58 internal nodes.
Tip labels:
Ranidae_ott364560, Rhacophoridae_ott432783, Mantellidae_ott38969, Dicroglossidae_ott1081208, Ranixalidae_ott403946, Nyctibatrachidae_ott1081210, ...
Node labels:
Amphibia ott544595, Batrachia ott471197, Anura ott991547, mrcaott114ott3129, mrcaott114ott37876, mrcaott114ott18818, ...
Rooted; no branch lengths.
ape::plot.phylo(amphibia_families_subtree, cex = 1.2)
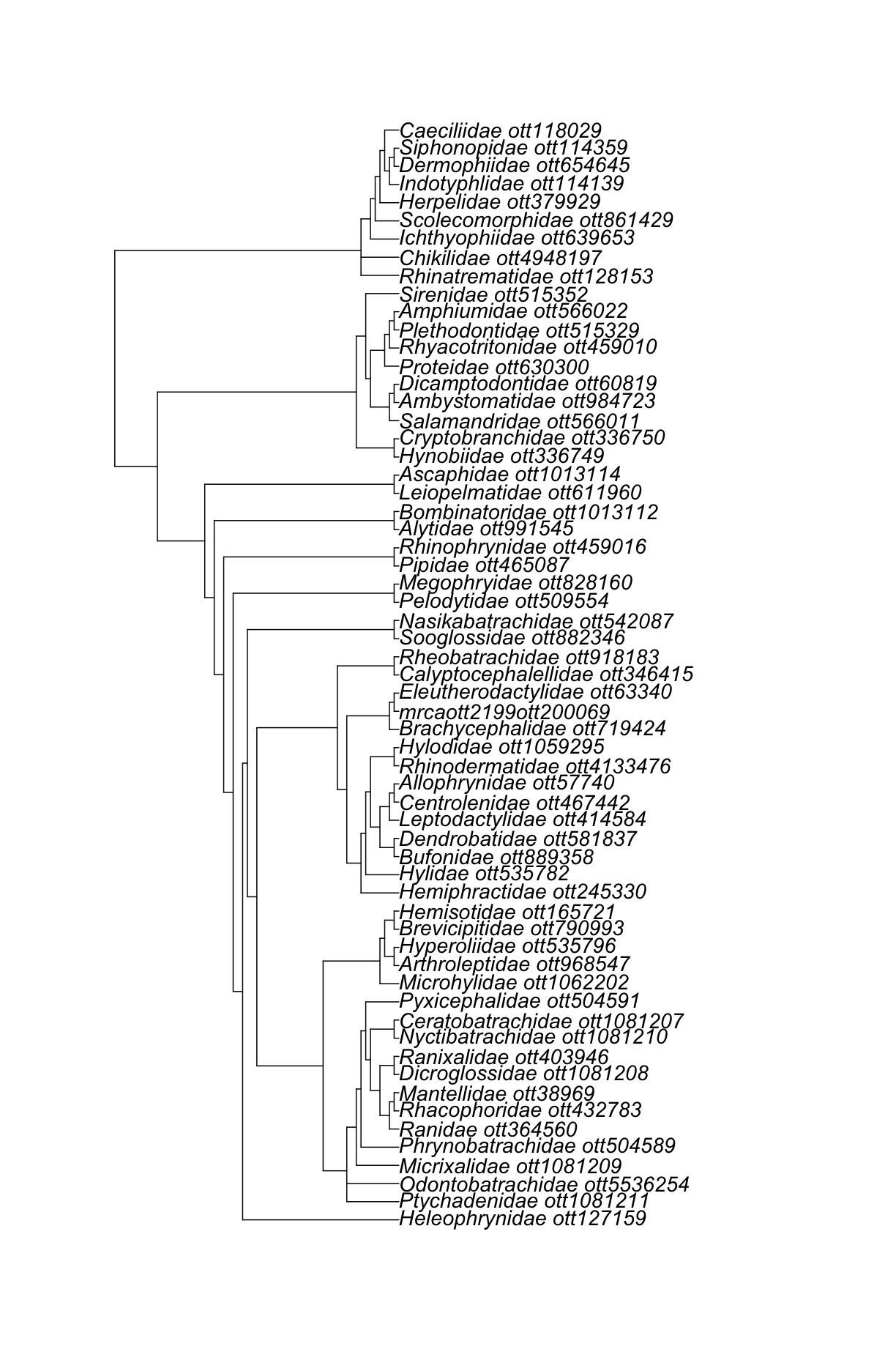 Super cool!
Super cool!
Hands on! Get a family subtree without ott ids in the tip labels
Hint: Look at the arguments of function
tol_induced_subtree()Solution
amphibia_families_subtree2 <- rotl::tol_induced_subtree(amphibia_families$Amphibia$ott_id, label_format = "name")Warning in collapse_singles(tr, show_progress): Dropping singleton nodes with labels: mrcaott114ott391676, mrcaott22019ott241869, mrcaott22583ott100573, mrcaott22583ott44382, mrcaott44382ott72638, mrcaott44382ott100564, mrcaott65695ott254163, mrcaott65695ott121259, mrcaott2199ott411156, mrcaott7464ott21502, mrcaott21502ott918196, Pelobatoidea, mrcaott18818ott47772, Sirenoidea, mrcaott154777ott464898, mrcaott464898ott540102ape::plot.phylo(amphibia_families_subtree2, cex = 1.2)
Hack: Get all taxa from a taxonomic rank.
There are several ways to do this using
rotlfunctions only. Try it out!Look at a possible solution
amphibia_taxonomy <- rotl::taxonomy_subtree(resolved_names["Amphibia",]$ott_id) ls(amphibia_taxonomy)[1] "edge_label" "tip_label"length(amphibia_taxonomy$tip_label)[1] 14156head(amphibia_taxonomy$tip_label)[1] "Caecilia_tentaculata_ott540102" "unclassified_Caecilia_ott7667087" [3] "Caecilia_gracilis_ott3617882" "Caecilia_volcani_ott923510" [5] "Caecilia_sp._ott9109" "Caeciliasp.KR-2007ott154780"tail(amphibia_taxonomy$tip_label)[1] "Aistopoda_ott5326022" [2] "Nectridea_ott5326000" [3] "Diadectomorpha_ott5326029" [4] "'Cheliderpeton(genusinDeuterostomia" [5] "'Sclerocephalus(genusinDeuterostomia" [6] "Ichthyostegalia_ott5326052"amphibia_taxonomy$edge_label[1] "'Caecilia(genusinDeuterostomia" [2] "ott9110'" [3] "Oscaecilia_ott654632" [4] "Mimosiphonops_ott3617932" [5] "Nectocaecilia_ott3617935" [6] "Sylvacaecilia_ott3617918" [7] "Parvicaecilia_ott3617941" [8] "Apodops_ott4132583" [9] "Caeciliidae_ott118029" [10] "Typhlonectes_ott639646" [11] "Chthonerpeton_ott633052" [12] "Atretochoana_ott3617929" [13] "Potamotyphlus_ott7667119" [14] "Typhlonectidae_ott639647" [15] "Ichthyophis_ott639652" [16] "Uraeotyphlus_ott498526" [17] "Caudacaecilia_ott831465" [18] "Bdellophis_ott4132629" [19] "Ichthyophiidae_ott639653" [20] "Dermophis_ott197731" [21] "Geotrypetes_ott283384" [22] "Schistometopum_ott568515" [23] "Gymnopis_ott80539" [24] "Dermophiidae_ott654645" [25] "Epicrionops_ott840081" [26] "Rhinatrema_ott498525" [27] "Rhinatrematidae_ott128153" [28] "Grandisonia_ott118032" [29] "Hypogeophis_ott118055" [30] "Gegeneophis_ott227124" [31] "Praslinia_ott80530" [32] "Indotyphlus_ott479403" [33] "Idiocranium_ott3617923" [34] "Indotyphlidae_ott114139" [35] "Microcaecilia_ott654633" [36] "Siphonops_ott80529" [37] "Brasilotyphlus_ott3617930" [38] "Luetkenotyphlus_ott479420" [39] "Siphonopidae_ott114359" [40] "Scolecomorphus_ott47815" [41] "Crotaphatrema_ott21512" [42] "Scolecomorphidae_ott861429" [43] "Boulengerula_ott42295" [44] "Herpele_ott77667" [45] "Herpelidae_ott379929" [46] "Chikila_ott5758907" [47] "Chikilidae_ott4948197" [48] "Wesserpeton_ott6159232" [49] "Albanerpeton_ott3617926" [50] "Anoualerpeton_ott3621326" [51] "Celtedens_ott3621303" [52] "Albanerpetontidae_ott3617924" [53] "Eocaecilia_ott4132637" [54] "Eocaeciliaidae_ott3617936" [55] "Rubricacaecilia_ott3617946" [56] "Gymnophiona_ott118027" [57] "Plethodon_ott515349" [58] "Karsenia_ott893551" [59] "Desmognathus_ott1050144" [60] "Ensatina_eschscholtzii_ott637537" [61] "Ensatina_ott64118" [62] "Aneides_ott64140" [63] "Hydromantes_no_rank_Hydromantes_ott693462" [64] "'Atylodes(genusinDeuterostomia" [65] "ott693464'" [66] "Hydromantes_imperialis_ott675243" [67] "Hydromantes_ambrosii_ott953902" [68] "Speleomantes_ott693468" [69] "Hydromantes_ott953899" [70] "Phaeognathus_ott964128" [71] "Plethodontinae_ott798661" [72] "Oeditriton_ott75459" [73] "Oedopinola_ott157731" [74] "Oedipina_subgenus_Oedipina_ott157732" [75] "Oedipina_ott99021" [76] "Cryptotriton_ott99024" [77] "Nototriton_ott99028" [78] "Dendrotriton_ott99034" [79] "Haideotriton_ott133635" [80] "Eladinea_ott3620958" [81] "Bolitoglossa_subgenus_Bolitoglossa_ott5544021" [82] "Nanotriton_ott4132647" [83] "Oaxakia_ott3620672" [84] "Mayamandra_ott3620674" [85] "Magnadigita_ott5544022" [86] "Pachymandra_ott3620664" [87] "Bolitoglossa_ott515331" [88] "Gyrinophilus_porphyriticus_ott1017696" [89] "Gyrinophilus_palleucus_ott790871" [90] "Gyrinophilus_ott1068026" [91] "Hemidactylium_ott798658" [92] "'Stereochilus(genusinOpisthokonta" [93] "ott798664'" [94] "Bradytriton_ott798666" [95] "Aquiloeurycea_ott5800410" [96] "Eurycea_troglodytes_complex_ott133631" [97] "Eurycea_multiplicata_ott839995" [98] "Eurycea_latitans_complex_ott839996" [99] "Eurycea_neotenes_complex_ott5265288" [100] "Eurycea_longicauda_ott534299" [101] "Eurycea_ott1023734" [102] "Parvimolge_ott46159" [103] "Ixalotriton_ott46161" [104] "Batrachoseps_ott64143" [105] "Urspelerpes_ott890499" [106] "Thorius_ott224304" [107] "Nyctanolis_ott224306" [108] "Chiropterotriton_ott224307" [109] "Isthmura_bellii_ott46162" [110] "Isthmura_sierraoccidentalis_ott98841" [111] "Isthmura_ott5800409" [112] "Pseudoeurycea_ott174965" [113] "Pseudotriton_montanus_ott401642" [114] "Pseudotriton_ruber_ott1017693" [115] "Pseudotriton_ott1017695" [116] "ott7666946'" [117] "Hemidactyliinae_ott149927" [118] "Hydromantoides_ott4132648" [119] "ott5336221'" [120] "Plethodontidae_ott515329" [121] "Necturus_ott963039" [122] "'Proteus(genusindomainEukaryota" [123] "ott666457'" [124] "Proteidae_ott630300" [125] "Dicamptodon_ott60824" [126] "Chrysotriton_ott4132676" [127] "Dicamptodontidae_ott60819" [128] "Rhyacotriton_ott459009" [129] "Rhyacotritonidae_ott459010" [130] "Lissotriton_boscai_ott830424" [131] "Lissotriton_vulgaris_ott1041774" [132] "Lissotriton_helveticus_ott9366" [133] "Lissotriton_ott389617" [134] "Paramesotriton_ott406723" [135] "Echinotriton_ott698358" [136] "Cynops_cyanurus_ott406736" [137] "Cynops_ott1041765" [138] "Taricha_torosa_ott953897" [139] "Taricha_ott1041792" [140] "Triturus_marmoratus_ott1041767" [141] "Triturus_karelinii_ott1041769" [142] "Triturus_dobrogicus_ott1041781" [143] "Triturus_carnifex_ott1041783" [144] "Triturus_ott1041799" [145] "Neurergus_strauchii_ott288025" [146] "Neurergus_ott288029" [147] "Ichthyosaura_alpestris_ott159060" [148] "Ichthyosaura_ott835314" [149] "Laotriton_ott4948201" [150] "Tylototriton_verrucosus_ott932561" [151] "Tylototriton_subgenus_Tylototriton_ott175813" [152] "Tylototriton_wenxianensis_ott981376" [153] "Yaotriton_ott175815" [154] "Tylototriton_ott456815" [155] "Pachytriton_ott499077" [156] "Notophthalmus_meridionalis_ott1031751" [157] "Notophthalmus_ott566013" [158] "Pleurodeles_waltl_ott566038" [159] "Pleurodeles_ott566021" [160] "Ommatotriton_ophryticus_ott645229" [161] "Ommatotriton_vittatus_ott865707" [162] "Ommatotriton_ott645233" [163] "Calotriton_ott793904" [164] "Euproctus_ott861431" [165] "Pleurodelinae_ott4948200" [166] "Salamandra_algira_ott308113" [167] "Salamandra_salamandra_ott689975" [168] "Salamandra_atra_ott689978" [169] "Salamandra_infraimmaculata_ott1024870" [170] "Salamandra_ott115714" [171] "Mertensiella_ott1066558" [172] "Chioglossa_ott499080" [173] "Lyciasalamandra_luschani_ott1012890" [174] "Lyciasalamandra_antalyana_ott1024875" [175] "Lyciasalamandra_fazilae_ott1024882" [176] "Lyciasalamandra_billae_ott1024889" [177] "Lyciasalamandra_ott800960" [178] "Salamandrinae_ott4948209" [179] "Salamandrina_ott674677" [180] "Salamandrininae_ott4948210" [181] "Pingia_ott4132654" [182] "Megalotriton_ott4132653" [183] "Bishara_ott4132664" [184] "Chelotriton_ott4132649" [185] "Oligosemia_ott3620637" [186] "Salamandridae_ott566011" [187] "Amphiuma_ott566025" [188] "Piceoerpeton_ott3620638" [189] "Paleoamphiuma_ott4132667" [190] "Proamphiuma_ott4132669" [191] "Amphiumidae_ott566022" [192] "'Ambystoma''unisexualhybridcomplex''ott203585'" [193] "Ambystoma_tigrinum_ott1092552" [194] "ott7071236'" [195] "Ambystoma_macrodactylum_ott938349" [196] "ott6159010'" [197] "Ambystoma_ott984720" [198] "Amblystoma_ott7666871" [199] "Ambystomatidae_ott984723" [200] "Salamandroidea_ott336747" [201] "Satobius_ott5800418" [202] "Poyarius_ott5800416" [203] "Hynobius_subgenus_Hynobius_ott5800417" [204] "Hynobius_ott953911" [205] "Pseudohynobius_ott1018812" [206] "Salamandrella_ott1021847" [207] "Pachyhynobius_ott1021854" [208] "Batrachuperus_ott101275" [209] "Iranodon_ott7071233" [210] "'Liua(genusinHolozoa" [211] "ott675465'" [212] "Ranodon_ott834698" [213] "Paradactylodon_ott838538" [214] "Onychodactylus_ott887098" [215] "Hynobiidae_ott336749" [216] "Cryptobranchus_alleganiensis_ott534288" [217] "Cryptobranchus_ott534314" [218] "Andrias_ott788372" [219] "Aviturus_ott3621219" [220] "Eoscapherpeton_ott3620822" [221] "Chunerpeton_ott3620515" [222] "Ulanurus_ott3620810" [223] "Cryptobranchidae_ott336750" [224] "Cryptobranchoidea_ott336753" [225] "Pseudobranchus_ott401981" [226] "Siren_intermedia_ott991553" [227] "Siren_ott991556" [228] "Kababisha_ott4132683" [229] "Habrosaurus_ott4132681" [230] "Sirenidae_ott515352" [231] "Sirenoidea_ott336754" [232] "Urupia_ott6376252" [233] "Regalerpeton_ott6376245" [234] "Nesovtriton_ott6376241" [235] "Pangerpeton_ott6376243" [236] "Seminobatrachus_ott6376247" [237] "Beiyanerpeton_ott6138079" [238] "Nasopus_ott7666998" [239] "Sinerpeton_ott3620651" [240] "Horezmia_ott3621060" [241] "Valdotriton_ott3620654" [242] "Jeholotriton_ott3620650" [243] "Kiyatriton_ott3620671" [244] "Liaoxitriton_ott3620675" [245] "Laccotriton_ott3620658" [246] "Iridotriton_ott3620676" [247] "Hemitrypus_ott3620678" [248] "Urodeles_ott7666996" [249] "Heteroclitotriton_ott3621355" [250] "Hylaeobatrachus_ott3620673" [251] "Marmorerpeton_ott3621285" [252] "Scapherpeton_ott4132706" [253] "Lisserpeton_ott4132711" [254] "Scapherpetontidae_ott3620677" [255] "Prodesmodon_ott4132716" [256] "Triassuridae_ott3620648" [257] "Noterpeton_ott4132718" [258] "Noterpetontidae_ott3620653" [259] "Karaurus_ott3620667" [260] "Kokartus_ott4132704" [261] "Karauridae_ott3620668" [262] "Opisthotriton_ott4132698" [263] "'Parrisia(genusinOpisthokonta" [264] "ott4132702'" [265] "Palaeoproteus_ott4132694" [266] "Batrachosauroides_ott4132696" [267] "Peratosauroides_ott4132695" [268] "Batrachosauroididae_ott3620655" [269] "Apricosiren_ott3620652" [270] "Ramonellus_ott4132714" [271] "Prosiren_ott4132713" [272] "Prosirenidae_ott3620660" [273] "Galverpeton_ott3620649" [274] "Caudata_ott984716" [275] "Allophryne_ott57742" [276] "Allophrynidae_ott57740" [277] "Celsiella_ott270773" [278] "Hyalinobatrachium_ott717620" [279] "Hyalinobatrachinae_ott1011638" [280] "Centrolene_ott9102" [281] "Nymphargus_ott467441" [282] "Cochranella_ott57743" [283] "Chimerella_ott434792" [284] "Rulyrana_ott511705" [285] "Teratohyla_ott270772" [286] "Espadarana_ott571380" [287] "Vitreorana_ott571381" [288] "Sachatamia_ott434800" [289] "Cochranellini_ott1011644" [290] "Centroleninae_ott1011645" [291] "Ikakogi_ott942704" [292] "Centrolenidae_ott467442" [293] "Allocentroleniae_ott9105" [294] "Flectonotus_ott580186" [295] "Stefania_ott787132" [296] "Gastrotheca_ott535780" [297] "Cryptobatrachus_ott1091968" [298] "Hemiphractus_ott655571" [299] "Fritziana_ott864397" [300] "Hemiphractidae_ott245330" [301] "Physalaemus_ott414579" [302] "Edalorhina_ott622247" [303] "Pleurodema_ott1032369" [304] "Pseudopaludicola_ott880717" [305] "Engystomops_ott1019408" [306] "Leiuperinae_ott216864" [307] "Lithodytes_ott315881" [308] "Hydrolaetare_ott791392" [309] "Adenomera_ott412180" [310] "Leptodactylus_gracilis_ott1045781" [311] "Leptodactylus_ott1036182" [312] "Leptodactylinae_ott4133298" [313] "Crossodactylodes_ott3620328" [314] "Rupirana_ott3620326" [315] "Scythrophrys_ott462991" [316] "Paratelmatobius_ott989417" [317] "Crossodactylodinae_ott4133299" [318] "ott5336507'" [319] "Arariphrynus_ott6158571" [320] "Physalalemus_ott6158772" [321] "Eorubeta_ott4133313" [322] "Eophractus_ott4133304" [323] "Niedenia_ott4133301" [324] "Indobatrachus_ott4133317" [325] "Neoprocoela_ott4133318" [326] "Phrynocerus_ott3619498" [327] "ott5336509'" [328] "Leptodactylidae_ott414584" [329] "Heleophryne_ott160485" [330] "Hadromophryne_ott971912" [331] "Heleophrynidae_ott127159" [332] "Proceratophrys_ott254172" [333] "Hylorina_ott440882" [334] "Alsodes_ott413976" [335] "Eupsophus_ott519523" [336] "Macrogenioglottus_ott726713" [337] "Odontophrynus_americanus_ott609773" [338] "Odontophrynus_ott1083887" [339] "Alsodidae_ott533096" [340] "Cycloramphus_ott254162" [341] "Zachaenus_ott346406" [342] "Thoropa_ott1008750" [343] "Limnomedusa_ott914517" [344] "Cycloramphidae_ott533097" [345] "Cruziohyla_ott329968" [346] "Agalychnis_ott407256" [347] "Pachymedusa_ott254792" [348] "Pithecopus_ott6158758" [349] "Phyllomedusa_ott364571" [350] "Phrynomedusa_ott528970" [351] "Hylomantis_ott1088100" [352] "Phasmahyla_ott1087177" [353] "Phyllomedusinae_ott245327" [354] "Nyctimystes_ott444690" [355] "Cyclorana_ott580206" [356] "Litoria_ott414588" [357] "Pelodryadinae_ott245328" [358] "Scarthyla_ott72615" [359] "Scinax_ott245332" [360] "Ololygon_ott6155586" [361] "Pseudis_ott1035865" [362] "Dendropsophus_ott206399" [363] "Xenohyla_ott510392" [364] "Sphaenorhynchus_ott1026159" [365] "Dendropsophini_ott678333" [366] "Hypsiboas_ott412457" [367] "Nesorohyla_ott7070190" [368] "Bokermannohyla_ott131345" [369] "Myersiohyla_ott206401" [370] "Hyloscirtus_ott131357" [371] "Boana_ott7070167" [372] "Aplastodiscus_ott570075" [373] "Cophomantini_ott678336" [374] "Osteopilus_ott59134" [375] "VG-2010ott590425'" [376] "Osteocephalus_ott95406" [377] "Itapotihyla_ott257368" [378] "Argenteohyla_siemersi_ott100569" [379] "Argenteohyla_ott578372" [380] "Corythomantis_ott442032" [381] "Nyctimantis_ott1087156" [382] "Tepuihyla_ott510385" [383] "'Phyllodytes(genusinDeuterostomia" [384] "ott185324'" [385] "Aparasphenodon_ott570084" [386] "Trachycephalus_ott291909" [387] "Lophiohylini_ott206403" [388] "Anotheca_ott59159" [389] "Megastomatohyla_ott433685" [390] "Tlalocohyla_ott257366" [391] "Charadrahyla_ott257371" [392] "Ecnomiohyla_ott433687" [393] "Exerodonta_ott257373" [394] "Isthmohyla_ott433695" [395] "Plectrohyla_ott263732" [396] "Ptychohyla_ott263741" [397] "Smilisca_ott315884" [398] "Acris_crepitans_ott59141" [399] "Acris_ott859275" [400] "Dryophytes_ott7070248" [401] "Pseudacris_crucifer_ott747287" [402] "Pseudacris_nigrita_ott80759" [403] "Pseudacris_ott173133" [404] "Triprion_ott238788" [405] "Bromeliohyla_ott729848" [406] "Sarcohyla_ott7070264" [407] "Duellmanohyla_ott148347" [408] "Hyla_annectans_ott655531" [409] "Hyla_arborea_ott677266" [410] "Hyla_ott1062216" [411] "Atlantihyla_ott7070220" [412] "Rheohyla_ott7070262" [413] "Quilticohyla_ott7070260" [414] "Hylini_ott131364" [415] "Hylinae_ott245329" [416] "Dryaderces_ott7666055" [417] "Diaglena_ott3620134" [418] "Proacris_ott4133404" [419] "Australobatrachus_ott3620437" [420] "ott5336513'" [421] "ott5336510'" [422] "Hylidae_ott535782" [423] "Yunganastes_ott558192" [424] "Tachiramantis_ott5810353" [425] "Pristimantis_ott911396" [426] "Oreobates_ott415886" [427] "Strabomantis_ott415907" [428] "Hypodactylus_ott427286" [429] "Lynchius_ott482785" [430] "Niceforonia_ott3620193" [431] "Phrynopus_ott646334" [432] "Strabomantinae_ott199517" [433] "Geobatrachus_ott3620197" [434] "Atopophrynus_ott3620196" [435] "Dischidodactylus_ott3620198" [436] "Strabomantidae_ott199520" [437] "Brachycephalus_ott719425" [438] "Ischnocnema_ott7461" [439] "Brachycephalidae_ott719424" [440] "Insuetophrynus_ott44381" [441] "Rhinoderma_ott566945" [442] "Rhinodermatidae_ott4133476" [443] "Rheobatrachus_ott918195" [444] "Rheobatrachidae_ott918183" [445] "Megaelosia_ott777178" [446] "Crossodactylus_ott777180" [447] "Hylodes_ott304507" [448] "Hylodidae_ott1059295" [449] "Diasporus_ott427881" [450] "Pelorius_ott570532" [451] "Euhyas_ott1091840" [452] "Eleutherodactylus_auriculatus_complex_ott296402" [453] "Eleutherodactylus_subgenus_Eleutherodactylus_ott514824" [454] "Syrrhophus_ott692789" [455] "Eleutherodactylus_ott889366" [456] "Eleutherodactylinae_ott478714" [457] "Phyzelaphryne_ott770925" [458] "Adelophryne_ott497500" [459] "Phyzelaphryninae_ott63339" [460] "Eleutherodactylidae_ott63340" [461] "Craugastor_ott1091841" [462] "Haddadus_ott45657" [463] "Craugastorinae_ott7069989" [464] "Ceuthomantis_ott411155" [465] "Ceuthomantinae_ott277741" [466] "Noblella_ott580143" [467] "Euparkerella_ott387681" [468] "Bryophryne_ott45674" [469] "Microkayla_ott7070002" [470] "Barycholos_ott442099" [471] "Psychrophrynella_ott682536" [472] "Holoaden_ott941607" [473] "Holoadeninae_ott446314" [474] "Craugastoridae_ott63341" [475] "Epipedobates_ott192968" [476] "Leucostethus_ott7070056" [477] "Ameerega_ott615037" [478] "Colostethus_ott840290" [479] "Silverstoneia_nubicola_ott638061" [480] "Silverstoneia_ott467431" [481] "Colostethinae_ott1096757" [482] "Andinobates_ott533200" [483] "Dendrobates_ott60809" [484] "Excidobates_ott169660" [485] "Phyllobates_ott266338" [486] "Ranitomeya_ott516331" [487] "Oophaga_ott271432" [488] "Minyobates_ott271434" [489] "Adelphobates_ott271436" [490] "Dendrobatinae_ott1096758" [491] "Hyloxalus_ott1096760" [492] "Hyloxalinae_ott1096759" [493] "Allobates_ott407145" [494] "Mannophryne_ott766335" [495] "Ectopoglossus_ott7070048" [496] "Aromobates_ott357382" [497] "Anomaloglossus_ott1059120" [498] "Rheobates_ott1059121" [499] "Prostherapis_ott4133407" [500] "Aromobatinae_ott1059126" [501] "Dendrobatidae_ott581837" [502] "Churamiti_ott104959" [503] "Didynamipus_ott152264" [504] "Incilius_ott284628" [505] "Ingerophrynus_ott284645" [506] "Amazophrynella_ott77765" [507] "Pelophryne_ott417709" [508] "Pedostibes_ott417714" [509] "Oreophrynella_ott317404" [510] "Blythophryne_ott5926045" [511] "Melanophryniscus_ott506359" [512] "Osornophryne_ott506361" [513] "Schismaderma_ott506368" [514] "Rentapia_ott7069968" [515] "Pseudobufo_ott3619739" [516] "Leptophryne_ott1000787" [517] "Nimbaphrynoides_ott618903" [518] "Rhinella_ott909159" [519] "Nectophryne_ott779708" [520] "Nectophrynoides_ott943451" [521] "Dendrophryniscus_ott943455" [522] "Bufoides_ott3619731" [523] "Strauchbufo_ott6158681" [524] "Sabahphrynus_ott4133471" [525] "'Werneria(genusinOpisthokonta" [526] "ott1039942'" [527] "Wolterstorffina_ott1039944" [528] "Vandijkophrynus_ott1058528" [529] "Phrynoidis_ott909163" [530] "Peltophryne_ott906146" [531] "Adenomus_ott206075" [532] "Ghatophryne_ott7069948" [533] "Altiphrynoides_ott3619745" [534] "Nannophryne_ott334613" [535] "Epidalea_ott334615" [536] "Anaxyrus_boreas_ott436373" [537] "Anaxyrus_americanus_ott889326" [538] "Anaxyrus_ott334620" [539] "Bufotes_pewzowi_ott1072351" [540] "Bufotes_boulengeri_ott312348" [541] "Bufotes_oblongus_ott501744" [542] "Bufotes_viridis_ott962317" [543] "Bufotes_ott334622" [544] "Stephopaedes_ott370320" [545] "Capensibufo_ott370330" [546] "Xanthophryne_ott534594" [547] "Sclerophrys_superciliaris_ott10751" [548] "Sclerophrys_ott284630" [549] "Barbarophryne_ott5800473" [550] "Parapelophryne_ott3619737" [551] "Frostius_ott776624" [552] "Mertensophryne_ott831481" [553] "Ansonia_ott884638" [554] "Atelopus_ott884664" [555] "Bufo_japonicus_ott889333" [556] "Bufo_gargarizans_ott279549" [557] "Bufo_ott187219" [558] "Sigalegalephrynus_ott7069984" [559] "Rhaebo_ott906145" [560] "Poyntonophrynus_ott334619" [561] "Crepidophryne_ott136609" [562] "Duttaphrynus_ott960164" [563] "Truebella_ott3619730" [564] "Metaphryniscus_ott3619751" [565] "Laurentophryne_ott3619734" [566] "Bufonidae_ott889358" [567] "Telmatobius_ott736944" [568] "Telmatobiinae_ott777187" [569] "Lepidobatrachus_ott414583" [570] "Ceratophrys_ott386111" [571] "Chacophrys_ott431943" [572] "Caudiverbera_ott1068325" [573] "Ceratophryinae_ott777188" [574] "Uberabatrachus_ott6158684" [575] "'Wawelia(genusinHolozoa" [576] "ott6158686'" [577] "Baurubatrachus_ott3620439" [578] "Ceratophryidae_ott1008932" [579] "Atelognathus_ott594313" [580] "Chaltenobatrachus_ott6158620" [581] "Batrachyla_ott636138" [582] "Batrachylidae_ott1008933" [583] "Hyloidea_ott535802" [584] "Conraua_ott532104" [585] "Petropedetes_ott780983" [586] "Hylarthroleptis_ott4133596" [587] "Petropedetidae_ott337155" [588] "Liurana_ott3620715" [589] "Liurananinae_ott5926192" [590] "Alcalus_ott5926145" [591] "Alcalinae_ott5926144" [592] "Cornufer_ott376748" [593] "Platymantis_ott376756" [594] "Ceratobatrachinae_ott5926147" [595] "Ceratobatrachidae_ott1081207" [596] "Micrixalus_ott1054404" [597] "Micrixalidae_ott1081209" [598] "Phylacomantis_ott155651" [599] "Vatomantis_ott155652" [600] "Duboimantis_ott155653" [601] "Laurentomantis_ott155654" [602] "Gephyromantis_subgenus_Gephyromantis_ott730430" [603] "Gephyromantis_ott65127" [604] "Guibemantis_subgenus_Guibemantis_ott189957" [605] "Pandanusicola_ott1087627" [606] "Guibemantis_ott65128" [607] "Blommersia_ott484428" [608] "Boehmantis_ott484429" [609] "Spinomantis_ott484431" [610] "Wakea_ott484449" [611] "bernhardi_group_ott189958" [612] "cowani_group_ott1087728" [613] "laevigata_group_ott189959" [614] "betsileo_group_ott189960" [615] "madagascariensis_group_ott189961" [616] "Mantella_ott38970" [617] "Ochthomantis_ott155642" [618] "Mantidactylus_subgenus_Mantidactylus_ott484430" [619] "Hylobatrachus_ott678996" [620] "Maitsomantis_ott678997" [621] "Brygoomantis_ott678998" [622] "Chonomantis_ott678999" [623] "Mantidactylus_ott283924" [624] "Tsingymantis_ott976861" [625] "Mantellinae_ott764198" [626] "Aglyptodactylus_ott858604" [627] "Laliostoma_ott1054409" [628] "Laliostominae_ott764199" [629] "Sahona_ott764195" [630] "Boophis_ulftunni_group_ott591709" [631] "Boophis_albipunctatus_group_ott764192" [632] "Boophis_albilabris_group_ott764193" [633] "Boophis_goudoti_group_ott764194" [634] "Boophis_luteus_group_ott291550" [635] "Boophis_microtympanum_group_ott291551" [636] "Boophis_majori_group_ott291552" [637] "Boophis_mandraka_group_ott291553" [638] "Boophis_rappiodes_group_ott291554" [639] "Boophis_no_rank_Boophis_ott764197" [640] "Boophis_ott750471" [641] "Boophinae_ott764200" [642] "Mantellidae_ott38969" [643] "Nyctibatrachus_ott579706" [644] "Lankanectes_ott1000557" [645] "Nyctibatrachidae_ott1081210" [646] "Ptychadena_ott799885" [647] "'Hildebrandtia(genusinOpisthokonta" [648] "ott805120'" [649] "Lanzarana_ott3618164" [650] "Ptychadenidae_ott1081211" [651] "Nidirana_ott7070883" [652] "Glandirana_ott407917" [653] "Meristogenys_ott447458" [654] "Pulchrana_ott5800479" [655] "Humerana_ott4133627" [656] "Sylvirana_ott5800480" [657] "Papurana_ott5800481" [658] "Abavorana_ott5800482" [659] "'Rugosa(genusinBilateria" [660] "ott5800477'" [661] "Hylarana_ott909265" [662] "Pelophylax_porosus_ott885925" [663] "Pelophylax_cf_ott7070897" [664] "'Pelophylaxsp.AP-2016ott5926238'" [665] "Pelophylax_ott915572" [666] "Sanguirana_ott939786" [667] "Amnirana_ott7070869" [668] "'Huia(genusinOpisthokonta" [669] "ott73241'" [670] "Amolops_ott594781" [671] "Sumaterana_ott7070942" [672] "Rana_temporaria_ott14718" [673] "Rana_macrocnemis_ott842743" [674] "Rana_arvalis_ott448372" [675] "Rana_aurora_ott771201" [676] "Rana_tagoi_ott887103" [677] "Rana_subgenus_Rana_ott6158879" [678] "Rana_sphenocephala_ott61437" [679] "Pantherana_ott390002" [680] "Trypheropsis_ott397283" [681] "Rana_clamitans_ott515378" [682] "Aquarana_ott457630" [683] "Zweifelia_ott517467" [684] "sp.RdS541ott514662'" [685] "Lithobates_ott194286" [686] "Rana_ott364550" [687] "Odorrana_ott440559" [688] "'Hydrophylax(genusinDeuterostomia" [689] "ott5800478'" [690] "Clinotarsus_ott196284" [691] "Indosylvirana_ott5800483" [692] "Staurois_ott491960" [693] "'Babina(genusinDeuterostomia" [694] "ott1075295'" [695] "Hemimantis_ott4133632" [696] "Pterorana_ott3618410" [697] "Phrynopsis_ott5336502" [698] "Montorana_ott4133633" [699] "Microdiscopus_ott4133623" [700] "ott5346643'" [701] "ott5336501'" [702] "Ranidae_ott364560" [703] "Sallywalkerana_ott7070960" [704] "Indirana_ott528405" [705] "Ranixalidae_ott403946" [706] "Frankixalus_ott5926244" [707] "Liuixalus_ott356004" [708] "Pseudophilautus_ott455940" [709] "Feihyla_palpebralis_ott905916" [710] "Feihyla_ott638687" [711] "Chiromantis_doriae_ott594714" [712] "Chiromantis_vittatus_ott389176" [713] "Chiromantis_ott750030" [714] "Philautus_ott858619" [715] "Gracixalus_ott940934" [716] "Kurixalus_ott1000416" [717] "Raorchestes_ott13294" [718] "Nyctixalus_ott140891" [719] "ott60920'" [720] "ott5820355'" [721] "'Rhacophorus(genusinDeuterostomia" [722] "ott174267'" [723] "Taruga_ott7071023" [724] "Beddomixalus_ott4133641" [725] "Mercurana_ott4133643" [726] "Nasutixalus_ott5926252" [727] "Theloderma_asperum_ott594756" [728] "Theloderma_ott899254" [729] "Polypedates_megacephalus_ott908329" [730] "Polypedates_leucomystax_species_complex_ott1016237" [731] "Polypedates_ott908335" [732] "Ghatixalus_ott715952" [733] "Rhacophorinae_ott223221" [734] "Buergeria_ott175633" [735] "Buergeriinae_ott223222" [736] "Indorana_ott7666775" [737] "Rhacophoridae_ott432783" [738] "Phrynobatrachus_calcaratus_species_complex_ott909610" [739] "Phrynobatrachus_natalensis_species_complex_ott128002" [740] "Phrynobatrachus_mababiensis_species_complex_ott255122" [741] "Phrynobatrachus_ott406527" [742] "Phrynobatrachidae_ott504589" [743] "Arthroleptella_ott274121" [744] "Strongylopus_ott41644" [745] "Poyntonia_ott475120" [746] "Anhydrophryne_ott113819" [747] "Cacosternum_nanum_ott676305" [748] "Cacosternum_ott155686" [749] "Tomopterna_ott376752" [750] "Microbatrachella_ott751405" [751] "Nothophryne_ott3618085" [752] "Ericabatrachus_ott3618087" [753] "Natalobatrachus_ott532110" [754] "Amietia_ott1093295" [755] "Cacosterninae_ott896265" [756] "Pyxicephalus_ott911281" [757] "Aubria_ott143511" [758] "Pyxicephalinae_ott98460" [759] "Pyxicephalidae_ott504591" [760] "Odontobatrachus_ott5541336" [761] "Odontobatrachidae_ott5536254" [762] "Occidozyga_ott841800" [763] "Ingerana_ott998828" [764] "Occidozyginae_ott98462" [765] "'Sphaerotheca(genusinHolozoa" [766] "ott340050'" [767] "Feirana_ott391260" [768] "Nanorana_ott88189" [769] "Chrysopaa_ott4133659" [770] "Limnonectes_kuhlii_species_complex_ott307539" [771] "Limnonectes_ott646946" [772] "Quasipaa_ott859097" [773] "Nannophrys_ott775806" [774] "Hoplobatrachus_ott214642" [775] "Euphlyctis_ott376757" [776] "Minervarya_ott7070717" [777] "Fejervarya_ott914765" [778] "Dicroglossinae_ott98463" [779] "Allopaa_ott4133656" [780] "Ombrana_ott4133654" [781] "Dicroglossidae_ott1081208" [782] "'Ranoidea(superfamilyinsuborderNeobatrachia" [783] "ott911276'" [784] "Spicospina_ott1039925" [785] "Uperoleia_ott940195" [786] "Crinia_ott414587" [787] "Pseudophryne_ott127182" [788] "Metacrinia_ott412471" [789] "Myobatrachus_ott919139" [790] "'Assa(genusinOpisthokonta" [791] "ott906692'" [792] "Geocrinia_ott21505" [793] "Paracrinia_ott989413" [794] "Arenophryne_ott442097" [795] "Taudactylus_ott1080055" [796] "Myobatrachinae_ott214195" [797] "Adelotus_ott276281" [798] "Megistolotis_ott276283" [799] "Heleioporus_ott412467" [800] "Mixophyes_ott1073180" [801] "Lechriodus_ott160126" [802] "Philoria_ott214194" [803] "Limnodynastes_dumerilii_ott799654" [804] "ott3620442'" [805] "Limnodynastes_ott336756" [806] "Notaden_ott660957" [807] "Neobatrachus_ott1086229" [808] "Limnodynastinae_ott737864" [809] "ott5336498'" [810] "Bryobatrachus_ott3620469" [811] "Myobatrachidae_ott940181" [812] "Telmatobufo_ott736941" [813] "Calyptocephallela_ott346416" [814] "Calyptocephalellidae_ott346415" [815] "Myobatrachoidea_ott408493" [816] "Nasikabatrachus_ott919154" [817] "Nasikabatrachidae_ott542087" [818] "Sechellophryne_ott81795" [819] "Sooglossus_ott462993" [820] "Sooglossidae_ott882346" [821] "Sooglossoidea_ott408494" [822] "Semnodactylus_ott62328" [823] "Morerella_ott85260" [824] "Cryptothylax_ott186193" [825] "Kassina_ott535794" [826] "Opisthothylax_ott645881" [827] "Afrixalus_paradorsalis_ott362841" [828] "Afrixalus_ott652142" [829] "'Tachycnemis(genusinDeuterostomia" [830] "ott750024'" [831] "Hyperolius_concolor_ott505134" [832] "Hyperolius_fusciventris_ott85268" [833] "Hyperolius_viridiflavus_ott652134" [834] "Hyperolius_nasutus_complex_ott1088069" [835] "Hyperolius_ocellatus_ott40418" [836] "Hyperolius_marmoratus_ott441372" [837] "Hyperolius_balfouri_ott3619048" [838] "Hyperolius_parallelus_ott3619054" [839] "Hyperolius_tuberculatus_complex_ott914494" [840] "Hyperolius_ott78177" [841] "Alexteroon_ott1018592" [842] "Phlyctimantis_ott462985" [843] "Acanthixalus_ott532108" [844] "Heterixalus_ott652126" [845] "Paracassina_ott3619071" [846] "Arlequinus_ott3619083" [847] "Chlorolius_ott3619081" [848] "Callixalus_ott3619073" [849] "Chrysobatrachus_ott3619077" [850] "Kassinula_ott3619075" [851] "Rappia_ott7666450" [852] "Tornierella_ott4133669" [853] "Pseudocassina_ott4133668" [854] "Hyperoliidae_ott535796" [855] "Hemisus_ott968536" [856] "Hemisotidae_ott165721" [857] "Spelaeophryne_ott660153" [858] "Breviceps_adspersus_ott3618690" [859] "Breviceps_ott811566" [860] "Probreviceps_macrodactylus_ott111206" [861] "Probreviceps_ott283931" [862] "Balebreviceps_ott348136" [863] "Callulina_ott51821" [864] "Brevicipitidae_ott790993" [865] "Otophryne_ott354857" [866] "Otophryninae_ott404351" [867] "Elachistocleis_ott186172" [868] "Dermatonotus_ott186184" [869] "Hypopachus_ott354846" [870] "Dasypops_ott513414" [871] "Relictivomer_ott190096" [872] "Myersiella_ott593737" [873] "Syncope_ott833322" [874] "Arcovomer_ott844415" [875] "Gastrophryne_olivacea_ott565409" [876] "Gastrophryne_ott1062221" [877] "Ctenophryne_ott21518" [878] "Hamptophryne_ott304796" [879] "Stereocyclops_ott258756" [880] "Chiasmocleis_ott930341" [881] "Gastrophryninae_ott404352" [882] "Mysticellus_ott7070618" [883] "'Phrynella(genusinOpisthokonta" [884] "ott401609'" [885] "Microhyla_ott1054402" [886] "Metaphrynella_ott127416" [887] "Uperodon_ott127425" [888] "Micryletta_ott304793" [889] "Ramanella_ott237282" [890] "Kaloula_conjuncta_ott492217" [891] "Kaloula_ott563549" [892] "Glyphoglossus_ott728826" [893] "Chaperina_ott1080078" [894] "Microhylinae_ott630299" [895] "Phrynomantis_ott828944" [896] "Phrynomerinae_ott630304" [897] "Adelastes_ott3618939" [898] "Adelastinae_ott5800508" [899] "Hoplophryne_ott111196" [900] "Melanobatrachus_ott701612" [901] "Melanobatrachinae_ott685276" [902] "Albericus_ott114125" [903] "Oxydactyla_ott635875" [904] "Austrochaperina_ott635878" [905] "Cophixalus_ott725335" [906] "Sphenophryne_ott918197" [907] "Aphantophryne_ott1018584" [908] "Liophryne_ott1039945" [909] "Genyophryne_ott21522" [910] "Copiula_ott442618" [911] "Oreophryne_ott968531" [912] "Choerophryne_ott594327" [913] "Genyophryninae_ott790988" [914] "Scaphiophryne_ott283929" [915] "Paradoxophyla_ott1091642" [916] "Scaphiophryninae_ott790989" [917] "Anodonthyla_ott352317" [918] "Rhombophryne_ott443908" [919] "Cophyla_ott474124" [920] "Stumpffia_ott538044" [921] "Anilany_ott5926118" [922] "Madecassophryne_ott3618941" [923] "Platypelis_ott828937" [924] "Plethodontohyla_ott828948" [925] "Cophylinae_ott790990" [926] "Dyscophus_ott155674" [927] "Calluella_ott442106" [928] "Dyscophinae_ott790991" [929] "Hylophorbus_ott178305" [930] "Gastrophrynoides_ott401602" [931] "Xenorhina_ott635867" [932] "Vietnamophryne_ott7070471" [933] "Pherohapsis_ott27649" [934] "Paedophryne_ott27667" [935] "Pseudocallulops_ott266512" [936] "Metamagnusia_ott266513" [937] "Barygenys_ott343024" [938] "Siamophryne_ott7070469" [939] "Callulops_ott442619" [940] "Mantophryne_ott562905" [941] "Oninia_ott789802" [942] "Asterophrys_ott930335" [943] "Asterophryinae_ott790992" [944] "Kalophrynus_ott595012" [945] "Kalophrynidae_ott977735" [946] "Melanophryne_ott484356" [947] "Synapturanus_ott1039919" [948] "Caluella_ott7666463" [949] "Parhoplophryne_ott3618937" [950] "Phrynocara_ott4133706" [951] "ott5336500'" [952] "Microhylidae_ott1062202" [953] "Scotobleps_ott406460" [954] "Nyctibates_ott475155" [955] "Leptodactylodon_ott487657" [956] "Astylosternus_ott502427" [957] "Leptopelis_ott750017" [958] "Cardioglossa_ott1028305" [959] "Trichobatrachus_ott780979" [960] "Arthroleptis_poecilonotus_ott257572" [961] "sp.ott968549'" [962] "Arthroleptis_ott968551" [963] "Pararthroleptis_ott4133674" [964] "Arthroleptidae_ott968547" [965] "Microhyloidea_ott1062201" [966] "Neobatrachia_ott535804" [967] "Leiopelma_ott611959" [968] "Vieraella_ott3620429" [969] "Leiopelmatidae_ott611960" [970] "Ascaphus_ott485832" [971] "Ascaphidae_ott1013114" [972] "Pelodytes_ott509553" [973] "Aerugoamnis_ott7666850" [974] "Tephrodytes_ott4132928" [975] "Miopelodytes_ott4132929" [976] "Pelodytidae_ott509554" [977] "Xenophrys_ott525145" [978] "Leptobrachium_liui_ott125247" [979] "Leptobrachium_ott828157" [980] "ott5800427'" [981] "ott5800430'" [982] "ott5800433'" [983] "ott5800425'" [984] "ott5800432'" [985] "ott5800431'" [986] "Leptolalax_ott828161" [987] "Brachytarsophrys_ott1026957" [988] "Leptobrachella_ott3618031" [989] "Oreolalax_ott125264" [990] "Megophrys_ott626504" [991] "Vibrissaphora_ott535112" [992] "'Scutiger(genusinHolozoa" [993] "ott542861'" [994] "Atympanophrys_ott542885" [995] "Ophryophryne_ott854980" [996] "Leptobatrachium_ott7666821" [997] "Megophryidae_ott828160" [998] "Pelobates_syriacus_ott1026962" [999] "Pelobates_fuscus_ott233459" [1000] "Pelobates_balcanicus_ott7071200" [1001] "Pelobates_ott703766" [1002] "Spea_ott1013103" [1003] "Scaphiopus_ott485826" [1004] "Prospea_ott6158980" [1005] "Elkobatrachus_ott6158976" [1006] "Macropelobates_ott4132922" [1007] "Eopelobates_ott4132923" [1008] "Pelobatidae_ott485821" [1009] "Pelobatoidea_ott485820" [1010] "Barbourula_ott95655" [1011] "Bombina_variegata_ott991539" [1012] "Bombina_ott991561" [1013] "Eobarbourula_ott7665789" [1014] "Bombinatoridae_ott1013112" [1015] "Alytes_obstetricans_ott118036" [1016] "Alytes_ott118035" [1017] "Alytinae_ott5334814" [1018] "Latonia_ott4948216" [1019] "Discoglossus_galganoi_ott461369" [1020] "Discoglossus_pictus_ott465082" [1021] "Discoglossus_ott465081" [1022] "Discoglossinae_ott5334815" [1023] "Bakonybatrachus_ott6158557" [1024] "Zaphrissa_ott4133723" [1025] "Saevesoederberghia_ott4133718" [1026] "Scotiophryne_ott4133710" [1027] "Procerobatrachus_ott4133722" [1028] "Prodiscoglossus_ott4133715" [1029] "Enneabatrachus_ott4133713" [1030] "Paradiscoglossus_ott4133707" [1031] "Estesina_ott4133709" [1032] "Paralatonia_ott3620479" [1033] "Aralobatrachus_ott3620756" [1034] "Kizylkuma_ott3621137" [1035] "Eodiscoglossus_ott3620474" [1036] "ott5334816'" [1037] "Alytidae_ott991545" [1038] "Rhinophrynus_ott459015" [1039] "Rhadinosteus_ott4133726" [1040] "Chelomophrynus_ott4133728" [1041] "Eorhinophrynus_ott4133724" [1042] "Rhinophrynidae_ott459016" [1043] "Hymenochirus_ott940172" [1044] "Pseudhymenochirus_ott140873" [1045] "Pipa_ott233481" [1046] "Pipinae_ott436357" [1047] "sp.BOLD:AAH0940ott37892'" [1048] "Silurana_ott940175" [1049] "Xenopus_laevis_ott465096" [1050] "Xenopus_subgenus_Xenopus_ott1000363" [1051] "'Xenopus(genusinDeuterostomia" [1052] "ott465090'" [1053] "Xenopodinae_ott940173" [1054] "Kuruleufenia_ott7666858" [1055] "Shomronella_ott4133741" [1056] "Oumtkoutia_ott4133743" [1057] "Saltenia_ott4133745" [1058] "Eoxenopoides_ott4133746" [1059] "Llankibatrachus_ott3617963" [1060] "Pachycentrata_ott3617959" [1061] "Shelania_ott4948223" [1062] "Pipidae_ott465087" [1063] "Cordicephalus_ott4948379" [1064] "Pipoidea_ott1013102" [1065] "Iberobatrachus_ott6158601" [1066] "Eurycephalella_ott6158580" [1067] "Cratia_ott6158577" [1068] "Gracilibatrachus_ott6158593" [1069] "Hungarobatrachus_ott6158599" [1070] "Avitabatrachus_ott3620464" [1071] "Prosalirus_ott3620489" [1072] "Thoraciliacus_ott3620434" [1073] "Aygroua_ott3620490" [1074] "Sunnybatrachus_ott3620477" [1075] "Estesiella_ott3620480" [1076] "Neusibatrachus_ott4133754" [1077] "Albionbatrachus_ott4133752" [1078] "Palaeobatrachus_ott3620432" [1079] "Palaeobatrachidae_ott3620433" [1080] "Gobiatoides_ott4133749" [1081] "Cretasalia_ott4133747" [1082] "Gobiates_ott4948381" [1083] "Gobiatidae_ott3620484" [1084] "Nezpercius_ott3620485" [1085] "'Ranoidea(genusinfamilyPelodryadidae" [1086] "ott7666856'" [1087] "Pelodryadidae_ott3620482" [1088] "Liaobatrachus_ott3620450" [1089] "Comobatrachus_ott3620457" [1090] "Theatonius_ott4133750" [1091] "Tregobatrachidae_ott3620458" [1092] "Hatzegobatrachus_ott3620471" [1093] "Vulcanobatrachus_ott3620454" [1094] "Beelzebufo_ott3620436" [1095] "Czatkobatrachus_ott3620448" [1096] "Anura_ott991547" [1097] "Batrachia_ott471197" [1098] "Altenglanerpeton_ott6137829" [1099] "Warrenisuchus_ott6376283" [1100] "Kirktonecta_ott6372803" [1101] "Oestocephalus_ott3620786" [1102] "Oestocephalidae_ott6373116" [1103] "Amphibiopodiscus_ott6137950" [1104] "Glanochthon_ott6372584" [1105] "Antarctosuchus_ott6137976" [1106] "Proxilodon_ott7667189" [1107] "Temnocorpichnus_ott7667208" [1108] "Huskerpeton_ott7667122" [1109] "Arachana_ott7665764" [1110] "Hyloplesion_ott4948565" [1111] "Orthocosta_ott5332417" [1112] "Seeleya_ott4948566" [1113] "Microbrachis_ott4948567" [1114] "Microbrachidae_ott4948564" [1115] "Palaeoherpeton_ott4948575" [1116] "Pteroplax_ott4948569" [1117] "Calligenethlon_ott4948574" [1118] "Pholiderpeton_ott4948576" [1119] "Leptophractus_ott4948570" [1120] "Nummulosaurus_ott3621136" [1121] "Aversor_ott3620936" [1122] "Eogyrinidae_ott4948568" [1123] "'Archeria(genusinOpisthokonta" [1124] "ott4948498'" [1125] "Cricotus_ott4948503" [1126] "Neopteroplax_ott4948500" [1127] "Archeriidae_ott4948497" [1128] "Anthracosaurus_ott4948495" [1129] "Eobaphetes_ott3620825" [1130] "Anthracosauridae_ott4948494" [1131] "Limnodytes_ott7667134" [1132] "Cyclotosaurus_ott4948420" [1133] "Capitosauroides_ott4948417" [1134] "Paracyclotosaurus_ott4948414" [1135] "ott5332412'" [1136] "Capitosauridae_ott4948413" [1137] "Osteophorus_ott5332413" [1138] "Eryops_ott4948428" [1139] "Actinodon_ott4948432" [1140] "Clamorosaurus_ott4948431" [1141] "Onchiodon_ott4948433" [1142] "Glaukerpeton_ott3621251" [1143] "Eryopidae_ott4948427" [1144] "'Plagiosaurus(genusinDeuterostomia" [1145] "ott3396668'" [1146] "Melanopelta_ott5332407" [1147] "Plagiosternum_ott4948437" [1148] "Gerrothorax_ott4948439" [1149] "Plagiobatrachus_ott4948438" [1150] "Plagiorophus_ott4948436" [1151] "Plagiosuchus_ott4948440" [1152] "Plagioscutum_ott3620942" [1153] "Aranetsia_ott3620938" [1154] "Plagiosauridae_ott4948435" [1155] "Hyperokynodon_ott6159417" [1156] "Tirraturhinus_ott6159427" [1157] "Trematosaurus_ott4948448" [1158] "Trematolestes_ott6159429" [1159] "Inflectosaurus_ott4948445" [1160] "Cosgriffius_ott3621074" [1161] "Microposaurus_ott4948451" [1162] "Aphaneramma_ott4948452" [1163] "Trematosuchus_ott4948455" [1164] "Platystega_ott4948456" [1165] "Erythrobatrachus_ott4948457" [1166] "Tertremoides_ott4948447" [1167] "Tertrema_ott4948446" [1168] "Stoschiosaurus_ott4948442" [1169] "Bukobaja_ott3620689" [1170] "Wantzosaurus_ott4948443" [1171] "Trematosauridae_ott4948441" [1172] "Derwentia_ott5332402" [1173] "Pneumatostega_ott4948591" [1174] "Trucheosaurus_ott4948590" [1175] "Laidleria_ott4948595" [1176] "Deltasaurus_ott4948594" [1177] "Rhytidosteus_ott4948593" [1178] "Peltostega_ott4948592" [1179] "Boreopelta_ott4948589" [1180] "Nanolania_ott3620978" [1181] "Sangaia_ott6159395" [1182] "Rhytidosteidae_ott4948586" [1183] "Pasawioops_ott6158525" [1184] "Rubeostratilia_ott6158529" [1185] "Gerobatrachus_ott6158519" [1186] "'Micropholis(genusinOpisthokonta" [1187] "ott5332409'" [1188] "Plemmyradytes_ott4948600" [1189] "Eoscopus_ott4948602" [1190] "Platyrhinops_ott4948601" [1191] "Amphibamus_ott4948603" [1192] "Georgenthalia_ott4948604" [1193] "Micromelerpeton_ott4948599" [1194] "Doleserpeton_ott4948597" [1195] "Tersomius_ott4948606" [1196] "Amphibamidae_ott4948596" [1197] "Platycepsion_ott6159073" [1198] "Banksiops_ott4948623" [1199] "Sinobrachyops_ott4948622" [1200] "Gobiops_ott3621200" [1201] "Bathignathus_ott3620690" [1202] "Vigilius_ott3621075" [1203] "Bothriceps_ott4948611" [1204] "Xenobrachyops_ott4948614" [1205] "Brachyops_ott4948617" [1206] "Batrachosuchus_ott4948618" [1207] "Hadrokkosaurus_ott4948616" [1208] "Vanastega_ott3620909" [1209] "Notobrachyops_ott4948621" [1210] "Brachyopidae_ott4948609" [1211] "Adamanterpeton_ott4948584" [1212] "Chenoprosopus_ott4948581" [1213] "Macrerpeton_ott4948580" [1214] "Cochleosaurus_ott4948579" [1215] "Procochleosaurus_ott4948578" [1216] "Nigerpeton_ott4145928" [1217] "Cochleosauridae_ott4948577" [1218] "Callistomordax_ott6138163" [1219] "Dutuitosaurus_ott7667152" [1220] "Metoposaurus_ott7667153" [1221] "Calamops_ott4948542" [1222] "Koskinonodon_ott3621174" [1223] "Apachesaurus_ott4948546" [1224] "ott4948553'" [1225] "Metoposauridae_ott4948540" [1226] "Broomistega_ott3621325" [1227] "Rhinesuchoides_ott4948557" [1228] "Rhinesuchus_ott4948562" [1229] "Australerpeton_ott4948561" [1230] "Uranocentrodon_ott4948563" [1231] "Laccosaurus_ott3620886" [1232] "Rhineceps_ott4948560" [1233] "Rhinesuchidae_ott4948555" [1234] "Trimerorhachis_ott4948628" [1235] "Dawsonerpeton_ott4948631" [1236] "Doragnathus_ott4948632" [1237] "Neldasaurus_ott4948633" [1238] "Lafonius_ott4948627" [1239] "ott5332408'" [1240] "Trimerorhachidae_ott4948624" [1241] "Koinia_ott3621294" [1242] "Uralosuchus_ott3621295" [1243] "Archegosaurus_ott4948521" [1244] "Tryphosuchus_ott4948529" [1245] "Collidosuchus_ott4948531" [1246] "Bageherpeton_ott4948532" [1247] "Bashkirosaurus_ott4948533" [1248] "Prionosuchus_ott4948526" [1249] "Konzhukovia_ott4948527" [1250] "Platyoposaurus_ott4948534" [1251] "Kashmirosaurus_ott3621235" [1252] "ott5326030'" [1253] "Archegosauridae_ott4948520" [1254] "Milnererpeton_ott3621379" [1255] "Schoenfelderpeton_ott4948489" [1256] "Branchiosaurus_ott4948490" [1257] "Leptorophus_ott3620998" [1258] "Tungussogyrinus_ott3621023" [1259] "Apateon_ott6148513" [1260] "Branchiosauridae_ott4948488" [1261] "Scapanops_ott6159192" [1262] "Aspidosaurus_ott5332410" [1263] "Zygosaurus_ott4948464" [1264] "Platyhystrix_ott4948462" [1265] "Brevidorsum_ott4948461" [1266] "Anakamacops_ott4948477" [1267] "Iratusaurus_ott4948459" [1268] "Dissorophus_ott4948465" [1269] "Eumicrerpeton_ott4948479" [1270] "Astreptorhachis_ott4948476" [1271] "Alegeinosaurus_ott4948475" [1272] "Broiliellus_ott4948471" [1273] "Conjunctio_ott4948470" [1274] "Kamacops_ott4948469" [1275] "Mazonerpeton_ott4948468" [1276] "Cacops_ott4948466" [1277] "Fayella_ott3621071" [1278] "Perryella_ott3620971" [1279] "Reiszerpeton_ott7667056" [1280] "Dissorophidae_ott4948458" [1281] "Nyranerpeton_ott7667163" [1282] "Eimerisaurus_ott4948483" [1283] "Branchierpeton_ott4948481" [1284] "Limnerpeton_ott4948484" [1285] "Limnerpeton_elegans_ott6159339" [1286] "Limnogyrinus_ott3620986" [1287] "Micromelerpetontidae_ott4948480" [1288] "Odenwaldia_ott4948408" [1289] "Thoosuchus_ott4948409" [1290] "Volgosuchus_ott4948410" [1291] "Benthosuchidae_ott4948405" [1292] "'Ranula(genusinDeuterostomia" [1293] "ott7667193'" [1294] "Seymouria_ott4948517" [1295] "Rhinosauriscus_ott4948512" [1296] "ott5332401'" [1297] "Seymouriidae_ott4948510" [1298] "Stegotretus_ott6375811" [1299] "Cardiocephalus_ott4948394" [1300] "Bolterpeton_ott3621080" [1301] "Elfridia_ott4948390" [1302] "Cymatorhiza_ott4948391" [1303] "Hylerpeton_ott4948393" [1304] "Pariotichus_ott4948399" [1305] "Leiocephalikon_ott4948397" [1306] "Euryodus_ott4948402" [1307] "Sparodus_ott4948404" [1308] "ott4948388'" [1309] "Gymnarthridae_ott4948387" [1310] "Tarsopterus_ott7667205" [1311] "Goniocephalus_ott6159217" [1312] "Palaeotriton_ott6159353" [1313] "Sillerpeton_ott3621204" [1314] "Phlegethontia_ott4948386" [1315] "ott5332415'" [1316] "Phlegethontiidae_ott4948385" [1317] "Ophiderpeton_ott5325979" [1318] "Coloraderpeton_ott4948384" [1319] "Ophiderpetontidae_ott4948383" [1320] "Luzocephalus_ott3620880" [1321] "Thenaropus_ott3620729" [1322] "Gnorhimosuchus_ott3620882" [1323] "Silvanerpeton_ott3620720" [1324] "Puertollanopus_ott4141858" [1325] "Pariostegus_ott3621359" [1326] "Allopus_ott3620919" [1327] "Iberospondylus_ott3621151" [1328] "Dendrerpeton_ott4133863" [1329] "Balanerpeton_ott3621046" [1330] "Eugyrinus_ott4133866" [1331] "Dendrysekos_ott3620759" [1332] "Dendrerpetontidae_ott3621047" [1333] "Slaugenhopia_ott3621279" [1334] "Tupilakosaurus_ott3621032" [1335] "Thabanchuia_ott3620977" [1336] "Batrachosuchoides_ott3621069" [1337] "Tupilakosauridae_ott3621033" [1338] "Eolydekkerina_ott3621374" [1339] "Edingerella_ott3621372" [1340] "Rewana_ott4133848" [1341] "Mahavisaurus_ott4133849" [1342] "Indobrachyops_ott4133850" [1343] "Indobrachyopidae_ott3620699" [1344] "Kuttycephalus_ott3621377" [1345] "Compsocerops_ott3621373" [1346] "Siderops_ott4133916" [1347] "Koolasuchus_ott4133914" [1348] "Pelorocephalus_ott4133913" [1349] "Keratobrachyops_ott3620693" [1350] "Chigutisauridae_ott3621378" [1351] "Deltaherpeton_ott6159127" [1352] "Greererpeton_ott3620905" [1353] "Colosteus_ott4133851" [1354] "Ichthyerpeton_ott3620724" [1355] "Colosteidae_ott3620902" [1356] "Quasicyclotosaurus_ott3620903" [1357] "Dvinosaurus_ott4133890" [1358] "Dvinosauridae_ott3621292" [1359] "Kourerpeton_ott4133893" [1360] "Kourerpetidae_ott3621296" [1361] "Parioxys_ott4133895" [1362] "Parioxyidae_ott3621297" [1363] "Melosaurus_ott4133889" [1364] "Melosauridae_ott3621077" [1365] "Rileymillerus_ott3621029" [1366] "Palatinerpeton_ott3621076" [1367] "Edops_ott4133888" [1368] "Edopidae_ott3621073" [1369] "Stenotosaurus_ott6159317" [1370] "Cherninia_ott3621376" [1371] "Jammerbergia_ott3620885" [1372] "Bulgosuchus_ott3621260" [1373] "Tatrasuchus_ott3621259" [1374] "Wetlugasaurus_ott4133840" [1375] "Wellesaurus_ott4133839" [1376] "Mastodonsaurus_ott3620723" [1377] "Archotosaurus_ott4133843" [1378] "Promastodonsaurus_ott4133845" [1379] "Eocyclotosaurus_ott4133846" [1380] "Kestrosaurus_ott4133847" [1381] "Xenotosuchus_ott3621222" [1382] "Komatosuchus_ott3621276" [1383] "Subcyclotosaurus_ott3620937" [1384] "Mastodonsauridae_ott3620685" [1385] "Rotaurisaurus_ott3620911" [1386] "Almasaurus_ott3620969" [1387] "Almasauridae_ott3620910" [1388] "Manubrantlia_ott3620907" [1389] "Otocratiidae_ott3621202" [1390] "Yarengiidae_ott3621318" [1391] "Yuanansuchus_ott4133911" [1392] "Heylerosauridae_ott3621317" [1393] "Tambachia_ott3621236" [1394] "Ecolsonia_ott4133912" [1395] "Mordex_ott3621361" [1396] "Phonerpeton_ott3621065" [1397] "Trematopsidae_ott3621321" [1398] "Fedexia_ott7667214" [1399] "Rotaryus_ott6159408" [1400] "Anconastes_ott4133792" [1401] "Acheloma_ott4133794" [1402] "Actiobates_ott4133791" [1403] "Trematopidae_ott3621234" [1404] "Isodectes_ott3621316" [1405] "Acroplous_ott4133873" [1406] "Erpetosaurus_ott4133872" [1407] "Eobrachyopidae_ott3620988" [1408] "Peltobatrachus_ott3621182" [1409] "Peltobatrachidae_ott3620962" [1410] "Intasuchus_ott4133902" [1411] "Syndyodosuchus_ott4133904" [1412] "Intasuchidae_ott3621040" [1413] "Sclerothorax_ott3620706" [1414] "Sclerothoracidae_ott3620707" [1415] "Moenkopisaurus_ott3620979" [1416] "Lydekkerina_ott3621100" [1417] "Deltacephalus_ott4133762" [1418] "Chomatobatrachus_ott4133764" [1419] "Lydekkerinidae_ott3620946" [1420] "Abiadisaurus_ott3620849" [1421] "Megalocephalus_ott3621381" [1422] "Kyrinion_ott3621293" [1423] "Baphetes_ott3620847" [1424] "Eucritta_ott3620850" [1425] "Loxomma_ott4133853" [1426] "Metaxygnathus_ott3621223" [1427] "Ventastega_ott3621221" [1428] "Baphetidae_ott3620848" [1429] "Caerorhachis_ott4133782" [1430] "Caerorhachidae_ott3621220" [1431] "Zatrachys_ott3620883" [1432] "Stegops_ott4133906" [1433] "Dasyceps_ott4133781" [1434] "Zatrachydidae_ott3621022" [1435] "Eoserpeton_ott3620854" [1436] "Limnopus_ott3620855" [1437] "Erpetobrachium_ott3620862" [1438] "Bradymedusa_ott3620866" [1439] "Erierpeton_ott3620863" [1440] "Cursipes_ott3620809" [1441] "Thinopus_ott3620801" [1442] "Ammobatrachus_ott3621263" [1443] "Adelospondylus_ott4133887" [1444] "Adelogyrinus_ott4133886" [1445] "Dolichopareias_ott4133885" [1446] "Palaeomolgophis_ott3621280" [1447] "Adelogyrinidae_ott3621262" [1448] "Silvadectes_ott7667035" [1449] "Oradectes_ott7667034" [1450] "Orobates_ott6159155" [1451] "Ambedus_ott4133825" [1452] "Phanerosaurus_ott4133824" [1453] "Stephanospondylus_ott4133823" [1454] "Diadectes_ott4133816" [1455] "Diadectidae_ott3621190" [1456] "Syphonodon_ott3621101" [1457] "Lysipterygium_ott3621049" [1458] "Pseudophlegethontia_ott4133854" [1459] "Pseudophlegethontiidae_ott3620959" [1460] "Lethiscus_ott4133856" [1461] "Lethiscidae_ott3620960" [1462] "Barillopus_ott3620989" [1463] "Eosaurus_ott3620995" [1464] "Iliodiscus_ott3620791" [1465] "Itemirella_ott3620746" [1466] "Pseudobradypus_ott3620744" [1467] "Matthewichnus_ott3620871" [1468] "Nectes_ott3620821" [1469] "Metaeus_ott3620700" [1470] "Hylopus_ott3620820" [1471] "Octopodichnus_ott3621170" [1472] "Eldeceeon_ott3621036" [1473] "Bruktererpeton_ott4133901" [1474] "Eusauropleura_ott4133899" [1475] "Gephyrostegus_ott4133896" [1476] "Gephyrostegidae_ott3621038" [1477] "Eoherpeton_ott4133775" [1478] "Eoherpetontidae_ott3621161" [1479] "Waggoneria_ott4133776" [1480] "Waggoneriidae_ott3621160" [1481] "Tseajaia_ott4133766" [1482] "Tseajaiidae_ott3621159" [1483] "Madygenerpeton_ott6159117" [1484] "Jarilinus_ott3621035" [1485] "Chroniosaurus_ott4133768" [1486] "Phratochronis_ott4133773" [1487] "Uralerpeton_ott4133769" [1488] "Suchonica_ott3620846" [1489] "Chroniosuchus_ott3620802" [1490] "Chroniosuchidae_ott3620803" [1491] "Timanosaurus_ott3620844" [1492] "Mauchchunkia_ott4133808" [1493] "Papposaurus_ott4133807" [1494] "Proterogyrinus_ott4133805" [1495] "Proterogyrinidae_ott3620841" [1496] "Solenodonsaurus_bohemicus_ott7667200" [1497] "Solenodonsaurus_ott4133803" [1498] "Solenodonsauridae_ott3620845" [1499] "Tokosauridae_ott3621320" [1500] "Jiyuanitectum_ott6159094" [1501] "Bystrowiella_ott6159090" [1502] "Yumenerpeton_ott7667022" [1503] "Dromotectum_ott3620985" [1504] "Axitectum_ott4133785" [1505] "Synesuchus_ott3620717" [1506] "Vyushkoviana_ott7667020" [1507] "Bystrowianidae_ott3620767" [1508] "Tulerpeton_ott3621243" [1509] "Tulerpetontidae_ott3621242" [1510] "Pholidogaster_ott3620765" [1511] "Diplovertebron_ott3620963" [1512] "Pholidogasteridae_ott3620766" [1513] "Cricotillus_ott4133783" [1514] "Cricotidae_ott3620764" [1515] "Emeroleter_ott4133834" [1516] "Bashkyroleter_ott4133836" [1517] "Macroleter_ott4133831" [1518] "Nycteroleter_ott4133828" [1519] "Rhipaeosaurus_ott4128674" [1520] "Nycteroleteridae_ott3620741" [1521] "Crassigyrinus_ott3621209" [1522] "Crassigyrinidae_ott3621210" [1523] "Ymeria_ott6159265" [1524] "Ichthyostega_ott3620842" [1525] "Ichthyostegidae_ott3620843" [1526] "Hynerpeton_ott3621155" [1527] "Whatcheeria_ott3621156" [1528] "Ossinodus_ott3621274" [1529] "Pederpes_ott3621152" [1530] "Occidens_ott3621187" [1531] "Whatcheeriidae_ott3621153" [1532] "Utaherpeton_ott3621157" [1533] "Baropezia_ott3621158" [1534] "Tridentichnus_ott3621090" [1535] "Megapezia_ott3621096" [1536] "Westlothiana_ott3621363" [1537] "Saxonerpeton_ott4133755" [1538] "Hapsidopareion_ott4133756" [1539] "Llistrofus_ott4133758" [1540] "Hapsidopareiontidae_ott3621365" [1541] "Asaphestera_ott3621342" [1542] "Crinodon_ott4133882" [1543] "Tuditanus_ott4133883" [1544] "Boii_ott4133884" [1545] "Tuditanidae_ott3621044" [1546] "Quasicaecilia_ott3620876" [1547] "Batropetes_ott4133868" [1548] "Carrolla_ott4133870" [1549] "Brachystelechidae_ott3621082" [1550] "Odonterpeton_ott4133871" [1551] "Odonterpetontidae_ott3621079" [1552] "Pantylus_ott3621084" [1553] "Trachystegos_ott4133867" [1554] "Pantylidae_ott3621083" [1555] "Tambaroter_ott6159351" [1556] "Ostodolepis_ott4133799" [1557] "Micraroter_ott4133801" [1558] "Pelodosotis_ott4133797" [1559] "Nannaroter_ott3621350" [1560] "Ostodolepidae_ott3620772" [1561] "Trihecaton_ott4133796" [1562] "Trihecatontidae_ott3620771" [1563] "Rhynchonkos_ott4133918" [1564] "Rhynchonkidae_ott3621346" [1565] "Paramicrobrachis_ott3621347" [1566] "Bissektia_ott3620703" [1567] "Collettosaurus_ott3620711" [1568] "Anomoeichnus_ott3620710" [1569] "Paleohelcura_ott3620709" [1570] "Batrachichnus_ott3620708" [1571] "Novascoticus_ott3621059" [1572] "Mesichnium_ott3621055" [1573] "Notalacerta_ott3620889" [1574] "Glossoliga_ott3620895" [1575] "Notamphibia_ott3620982" [1576] "Batrachiderpeton_ott3620973" [1577] "Batrachiderpetontidae_ott3620972" [1578] "Montcellia_ott6159455" [1579] "Ctenerpeton_ott4133874" [1580] "Ptyonius_ott4133876" [1581] "Sauropleura_ott3621323" [1582] "Crossotelos_ott4133879" [1583] "Urocordylus_ott4133877" [1584] "Lepterpeton_ott4133878" [1585] "Urocordylidae_ott3620984" [1586] "Keraterpeton_ott3621322" [1587] "Diceratosaurus_ott4133907" [1588] "Diplocaulus_ott3620731" [1589] "Keraterpetontidae_ott3620732" [1590] "Ductilodon_ott3620975" [1591] "Diploceraspis_ott3620858" [1592] "Diplocaulidae_ott3621232" [1593] "Karpinskiosaurus_ott4133790" [1594] "Bystrowiana_ott4133788" [1595] "Kotlassia_ott3621230" [1596] "Kotlassiidae_ott3621231" [1597] "Spinarerpeton_ott6159180" [1598] "Makowskia_ott7667039" [1599] "Ariekanerpeton_ott4133858" [1600] "Utegenia_ott4133859" [1601] "Letoverpeton_ott4133860" [1602] "Discosauriscus_ott3621277" [1603] "Melanerpeton_ott4133857" [1604] "Discosauriscidae_ott3621042" [1605] "Datheosaurus_ott3620928" [1606] "Diplopelma_ott3620726" [1607] "Stanocephalosaurus_ott3621227" [1608] "Crucipes_ott3620798" [1609] "Adenoderma_ott3620770" [1610] "Marpurgichnium_ott3621309" [1611] "Hyloidichnus_ott3621312" [1612] "Ceratophryne_ott3621314" [1613] "Agostopus_ott3620834" [1614] "Palaeosauropus_ott3620828" [1615] "Acherontiscus_ott3621089" [1616] "Acherontiscidae_ott3621088" [1617] "Arizonerpeton_ott3621250" [1618] "Nanopus_ott3621031" [1619] "Capetus_ott3621255" [1620] "Asperipes_ott3620762" [1621] "Dromillopus_ott3621304" [1622] "Spathicephalus_ott3621330" [1623] "Dromopus_ott3621138" [1624] "Pleuroptyx_ott3621124" [1625] "Brachydectes_ott6159058" [1626] "Cocytinidae_ott3621140" [1627] "Lysorocephalus_ott3621311" [1628] "Lysorophus_ott3620916" [1629] "Molgophis_ott4133778" [1630] "Lysorophidae_ott3620917" [1631] "Pomatops_ott3621238" [1632] "Nanipes_ott3621240" [1633] "Ferganobatrachus_ott3621067" [1634] "Lapillopsis_ott3621072" [1635] "Crenipes_ott3621064" [1636] "Strictipes_ott3620941" [1637] "Foliipes_ott3620968" [1638] "Acerastea_ott3620970" [1639] "Devipes_ott3620965" [1640] "Biarmica_ott3620964" [1641] "Acanthostega_ott3620966" [1642] "Acanthostegidae_ott3620967" [1643] "Acutipes_ott3620939" [1644] "Barypodus_ott3621120" [1645] "Ichnium_ott7667127" [1646] "Erpetosuchus_ott6149016" [1647] "Discodactylus_ott5338047" [1648] "Amphibiopus_ott4141536" [1649] "Lehahichnus_ott4144213" [1650] "Saharastega_ott4145925" [1651] "Uruyiella_ott4145931" [1652] "Laidleriidae_ott4145930" [1653] "Ruecklinichnium_ott7667197" [1654] "Triadobatrachus_ott4948636" [1655] "Miobatrachus_ott4948635" [1656] "Protobatrachidae_ott4948634" [1657] "Proanura_ott5325992" [1658] "Eobatrachus_ott7667064" [1659] "Triassurus_ott6159433" [1660] "ott5326050'" [1661] "ott5736797'" [1662] "ott5326010'" [1663] "ott4948486'" [1664] "ott5332403'" [1665] "'Actinodontidae(familyinDeuterostomia" [1666] "ott4948485'" [1667] "Temnospondyli_ott5325987" [1668] "Amphibia_ott544595"edges <- datelife::extract_ott_ids(x=amphibia_taxonomy$edge_label)After extracting ott ids, there are some non numeric elements:'Caecilia(genusinDeuterostomia ott9110' 'Atylodes(genusinDeuterostomia ott693464' 'Stereochilus(genusinOpisthokonta ott798664' ott7666946' ott5336221' 'Proteus(genusindomainEukaryota ott666457' 'Ambystoma''unisexualhybridcomplex''ott203585' ott7071236' ott6159010' 'Liua(genusinHolozoa ott675465' 'Parrisia(genusinOpisthokonta ott4132702' ott5336507' ott5336509' VG-2010ott590425' 'Phyllodytes(genusinDeuterostomia ott185324' ott5336513' ott5336510' 'Werneria(genusinOpisthokonta ott1039942' 'Wawelia(genusinHolozoa ott6158686' 'Hildebrandtia(genusinOpisthokonta ott805120' 'Rugosa(genusinBilateria ott5800477' 'Pelophylaxsp.AP-2016ott5926238' 'Huia(genusinOpisthokonta ott73241' sp.RdS541ott514662' 'Hydrophylax(genusinDeuterostomia ott5800478' 'Babina(genusinDeuterostomia ott1075295' ott5346643' ott5336501' ott60920' ott5820355' 'Rhacophorus(genusinDeuterostomia ott174267' 'Sphaerotheca(genusinHolozoa ott340050' 'Ranoidea(superfamilyinsuborderNeobatrachia ott911276' 'Assa(genusinOpisthokonta ott906692' ott3620442' ott5336498' 'Tachycnemis(genusinDeuterostomia ott750024' 'Phrynella(genusinOpisthokonta ott401609' ott5336500' sp.ott968549' ott5800427' ott5800430' ott5800433' ott5800425' ott5800432' ott5800431' 'Scutiger(genusinHolozoa ott542861' ott5334816' sp.BOLD:AAH0940ott37892' 'Xenopus(genusinDeuterostomia ott465090' 'Ranoidea(genusinfamilyPelodryadidae ott7666856' 'Archeria(genusinOpisthokonta ott4948498' ott5332412' 'Plagiosaurus(genusinDeuterostomia ott3396668' 'Micropholis(genusinOpisthokonta ott5332409' ott4948553' ott5332408' ott5326030' 'Ranula(genusinDeuterostomia ott7667193' ott5332401' ott4948388' ott5332415' ott5326050' ott5736797' ott5326010' ott4948486' ott5332403' 'Actinodontidae(familyinDeuterostomia ott4948485'NAs removed.length(edges)[1] 1572edges_taxon_info <- rotl::taxonomy_taxon_info(edges) ls(edges_taxon_info[[1]])[1] "flags" "is_suppressed" [3] "is_suppressed_from_synth" "name" [5] "ott_id" "rank" [7] "source" "synonyms" [9] "tax_sources" "unique_name"is_family <- unname(unlist(sapply(edges_taxon_info, "[", "rank") %in% "family")) is_suppressed <- unname(unlist(sapply(edges_taxon_info, "[", "is_suppressed_from_synth"))) # flag "is suppressed from synth" is not updated, so it is useless for now. amphibia_families <- unname(unlist(sapply(edges_taxon_info, "[", "ott_id")[is_family])) in_tree <- rotl::is_in_tree(amphibia_families) amphibia_families_subtree <- rotl::tol_induced_subtree(amphibia_families[in_tree])Warning in collapse_singles(tr, show_progress): Dropping singleton nodes with labels: mrcaott114ott391676, mrcaott22019ott241869, mrcaott22583ott100573, mrcaott22583ott44382, mrcaott44382ott72638, mrcaott44382ott100564, mrcaott65695ott254163, mrcaott65695ott121259, mrcaott2199ott411156, mrcaott2199ott22795, mrcaott7464ott21502, mrcaott21502ott918196, Pelobatoidea ott485820, mrcaott18818ott47772, Sirenoidea ott336754, mrcaott154777ott464898, mrcaott464898ott540102
We have seen up to now how to get a portion of the synthetic OTOL. How do I inspect the source phylogenetic trees that support the subtrees?
Key Points
It is possible to get all types of subsets from the synthetic tree, as long as you can get the OTT ids!
5. Getting studies and trees supporting relationships in a synthetic subtree
Overview
Teaching: 5 min
Exercises: 5 minQuestions
What are the original studies supporting relationships in my synthetic subtree?
Objectives
Get supporting trees for certain regions of the synthetic Open Tree of Life.
To get the source trees supporting a node from our synthetic tree we will need two functions.
The function source_list() gets the study and tree ids (and other info) from source studies (not the trees). It is applied to a ‘tol_node’ object.
We already have one that we generated with tol_node_info(), do you remember how we called it?
Hands on! Get all supporting trees.
Get the supporting study metadata from the Canis node info. Store it in an object called
canis_node_studies. Look at its class and the information it contains.canis_node_studies <- rotl::source_list(canis_node_info)class(canis_node_studies)[1] "data.frame"str(canis_node_studies)'data.frame': 5 obs. of 3 variables: $ study_id: chr "ot_278" "ot_328" "pg_1428" "pg_2647" ... $ tree_id : chr "tree1" "tree1" "tree2855" "tree6169" ... $ git_sha : chr "3008105691283414a18a6c8a728263b2aa8e7960" "3008105691283414a18a6c8a728263b2aa8e7960" "3008105691283414a18a6c8a728263b2aa8e7960" "3008105691283414a18a6c8a728263b2aa8e7960" ...
Now that we have the ids, we can use the function get_study_tree(), which will get us the actual supporting trees.
This function takes one study id and tree id at a time, like this:
x <- 1
rotl::get_study_tree(study_id = canis_node_studies$study_id[x], tree_id = canis_node_studies$tree_id[x], tip_label="ott_taxon_name", deduplicate = TRUE)
Warning: Some tip labels were duplicated and have been modified: Leptocyon,
Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Canidae,
Canidae, Urocyon, Urocyon, Urocyon, Cerdocyon, Canis, Canis, Canis, Canis,
Canis, Canis, Canis, Canis, Canis, Canidae, Cynarctoides
Phylogenetic tree with 142 tips and 141 internal nodes.
Tip labels:
Prohesperocyon_wilsoni, Ectopocynus_antiquus, Ectopocynus_intermedius, Ectopocynus_simplicidens, Hesperocyon, Hesperocyon_gregarius, ...
Rooted; includes branch lengths.
Hands on! Get all supporting trees.
Call the output canis_source_trees
Hint: You can use a “for” loop or an
apply()function to get them all.Solution
With a ‘for’ loop.
canis_source_trees <- vector(mode = "list") # generate an empty list for (i in seq(nrow(canis_node_studies))){ source_tree <- rotl::get_study_tree(study_id = canis_node_studies$study_id[i], tree_id = canis_node_studies$tree_id[i], tip_label="ott_taxon_name", deduplicate = TRUE) canis_source_trees <- c(canis_source_trees, list(source_tree)) }Warning: Some tip labels were duplicated and have been modified: Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Canidae, Canidae, Urocyon, Urocyon, Urocyon, Cerdocyon, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canidae, Cynarctoidescanis_source_trees[[1]] Phylogenetic tree with 142 tips and 141 internal nodes. Tip labels: Prohesperocyon_wilsoni, Ectopocynus_antiquus, Ectopocynus_intermedius, Ectopocynus_simplicidens, Hesperocyon, Hesperocyon_gregarius, ... Rooted; includes branch lengths. [[2]] Phylogenetic tree with 294 tips and 272 internal nodes. Tip labels: Homo_sapiens, Rattus_norvegicus, Mus_musculus, Artibeus_jamaicensis, Mystacina_tuberculata, Tadarida_brasiliensis, ... Rooted; includes branch lengths. [[3]] Phylogenetic tree with 169 tips and 168 internal nodes. Tip labels: Xenopus_laevis, Anolis_carolinensis, Gallus_gallus, Taeniopygia_guttata, Tachyglossus_aculeatus, Ornithorhynchus_anatinus, ... Rooted; includes branch lengths. [[4]] Phylogenetic tree with 86 tips and 85 internal nodes. Tip labels: *tip_#1_not_mapped_to_OTT._Original_label_-_Morganucodon_oehleri, *tip_#2_not_mapped_to_OTT._Original_label_-_Morganucodon_watsoni, *tip_#3_not_mapped_to_OTT._Original_label_-_Haldanodon_exspectatus, Eomaia_scansoria, Amblysomus_hottentotus, Echinops_telfairi, ... Rooted; no branch lengths. [[5]] Phylogenetic tree with 78 tips and 77 internal nodes. Tip labels: Ornithorhynchus, Manis, Ailuropoda, Canis, Felis, Panthera, ... Rooted; no branch lengths.With an
apply()function.canis_source_trees <- sapply(seq(nrow(canis_node_studies)), function(i) rotl::get_study_tree(study_id = canis_node_studies$study_id[i], tree_id = canis_node_studies$tree_id[i], tip_label="ott_taxon_name", deduplicate = TRUE))Warning: Some tip labels were duplicated and have been modified: Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Leptocyon, Canidae, Canidae, Urocyon, Urocyon, Urocyon, Cerdocyon, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canis, Canidae, Cynarctoidescanis_source_trees[[1]] Phylogenetic tree with 142 tips and 141 internal nodes. Tip labels: Prohesperocyon_wilsoni, Ectopocynus_antiquus, Ectopocynus_intermedius, Ectopocynus_simplicidens, Hesperocyon, Hesperocyon_gregarius, ... Rooted; includes branch lengths. [[2]] Phylogenetic tree with 294 tips and 272 internal nodes. Tip labels: Homo_sapiens, Rattus_norvegicus, Mus_musculus, Artibeus_jamaicensis, Mystacina_tuberculata, Tadarida_brasiliensis, ... Rooted; includes branch lengths. [[3]] Phylogenetic tree with 169 tips and 168 internal nodes. Tip labels: Xenopus_laevis, Anolis_carolinensis, Gallus_gallus, Taeniopygia_guttata, Tachyglossus_aculeatus, Ornithorhynchus_anatinus, ... Rooted; includes branch lengths. [[4]] Phylogenetic tree with 86 tips and 85 internal nodes. Tip labels: *tip_#1_not_mapped_to_OTT._Original_label_-_Morganucodon_oehleri, *tip_#2_not_mapped_to_OTT._Original_label_-_Morganucodon_watsoni, *tip_#3_not_mapped_to_OTT._Original_label_-_Haldanodon_exspectatus, Eomaia_scansoria, Amblysomus_hottentotus, Echinops_telfairi, ... Rooted; no branch lengths. [[5]] Phylogenetic tree with 78 tips and 77 internal nodes. Tip labels: Ornithorhynchus, Manis, Ailuropoda, Canis, Felis, Panthera, ... Rooted; no branch lengths.
The object canis_node_studies contains a lot of information. You can get it using a ‘for’ loop, or an apply() function.
A key piece of information are the citations from the supporting studies. We can get these for each source trees with the function get_study_meta(). Let’s do it. First we need the study meta:
canis_node_studies_meta <- lapply(seq(nrow(canis_node_studies)), function(i)
rotl::get_study_meta(study_id = canis_node_studies$study_id[i]))
Now we can get the citations:
canis_node_studies_citations <- sapply(seq(length(canis_node_studies_meta)), function (i) canis_node_studies_meta[[i]]$nexml$`^ot:studyPublicationReference`)
Finally, let’s plot the supporting trees along with their citations.
for (i in seq(length(canis_source_trees))){
print(paste("The supporting tree below has", length(canis_source_trees[[i]]$tip.label), "tips."))
print(paste("Citation is:", canis_node_studies_citations[i]))
ape::plot.phylo(canis_source_trees[[i]])
}
[1] "The supporting tree below has 142 tips."
[1] "Citation is: Tedford, Richard H.; Wang, Xiaoming; Taylor, Beryl E. (2009). Phylogenetic systematics of the North American fossil Caninae (Carnivora, Canidae). Bulletin of the American Museum of Natural History, no. 325. http://hdl.handle.net/2246/5999\n\nWang, Xiaoming; Tedford, Richard H.; Taylor, Beryl E. (1999). Phylogenetic systematics of the Borophaginae (Carnivora, Canidae). Bulletin of the American Museum of Natural History, no. 243. http://hdl.handle.net/2246/1588\n\nWang, Xiaoming (1994). Phylogenetic systematics of the Hesperocyoninae (Carnivora, Canidae). Bulletin of the American Museum of Natural History, no. 221. http://hdl.handle.net/2246/829\n"
[1] "The supporting tree below has 294 tips."
[1] "Citation is: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[1] "The supporting tree below has 169 tips."
[1] "Citation is: Meredith, R.W., Janecka J., Gatesy J., Ryder O.A., Fisher C., Teeling E., Goodbla A., Eizirik E., Simao T., Stadler T., Rabosky D., Honeycutt R., Flynn J., Ingram C., Steiner C., Williams T., Robinson T., Herrick A., Westerman M., Ayoub N., Springer M., & Murphy W. 2011. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science 334 (6055): 521-524."

[1] "The supporting tree below has 86 tips."
[1] "Citation is: O'Leary, M. A., J. I. Bloch, J. J. Flynn, T. J. Gaudin, A. Giallombardo, N. P. Giannini, S. L. Goldberg, B. P. Kraatz, Z.-X. Luo, J. Meng, X. Ni, M. J. Novacek, F. A. Perini, Z. S. Randall, G. W. Rougier, E. J. Sargis, M. T. Silcox, N. B. Simmons, M. Spaulding, P. M. Velazco, M. Weksler, J. R. Wible, A. L. Cirranello. 2013. The placental mammal ancestor and the post-K-Pg radiation of placentals. Science 339 (6120): 662-667."

[1] "The supporting tree below has 78 tips."
[1] "Citation is: Lartillot, Nicolas, Frédéric Delsuc. 2012. Joint reconstruction of divergence times and life-history evolution in placental mammals using a phylogenetic covariance model. Evolution 66 (6): 1773-1787."
Note that the supporting trees for a node can be larger than the subtree itself.
You will have to drop the unwanted taxa from the supporting studies if you just want the parts that belong to the subtree.
Moreover, the tip labels have different taxon names in the source trees and the synthetic subtrees. I you go to the browser, you can access original tips and matched tips, but R drops that info. We would have to standardize them with TNRS before trying to subset, and that takes some time and often visual inspection.
Key Points
Supporting trees usually contain more taxa than the ones we are interested in.
6. Getting branch length information (proportional to time) for you taxa
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I find supporting trees that include branch lengths?
How do I subset them to include just the taxa I am interested in?
Objectives
Learn about the
opentree_chronogramsobject from datelife.Get source chronograms from the
opentree_chronogramsobject for a set of taxa.
What if I want to search the OTOL database (phylesystem) for studies and trees matching some criteria?
The rotl package has functions that allow getting a list of studies or source trees matching specific criteria.
You will recognise these functions because they start with the word studies_.
Now, what kind of properties can we search for in the OTOL database?
The function studies_properties() gets for us two lists, one for studies and another one for tree properties available for search.
Take a look at them:
rotl::studies_properties()
$study_properties
[1] "dc:subject" "dc:date"
[3] "ot:messages" "dc:title"
[5] "skos:changeNote" "ot:studyPublicationReference"
[7] "ot:candidateTreeForSynthesis" "ot:taxonLinkPrefixes"
[9] "treebaseId" "ot:focalCladeOTTTaxonName"
[11] "prism:modificationDate" "dc:contributor"
[13] "dc:creator" "xmlns"
[15] "ot:curatorName" "prism:number"
[17] "tb:identifier.study.tb1" "id"
[19] "ot:otusElementOrder" "ot:dataDeposit"
[21] "skos:historyNote" "ot:treesElementOrder"
[23] "prism:endingPage" "prism:section"
[25] "nexml2json" "ot:notIntendedForSynthesis"
[27] "ntrees" "treesById"
[29] "about" "prism:publicationName"
[31] "tb:identifier.study" "ot:studyYear"
[33] "otusById" "nexmljson"
[35] "ot:annotationEvents" "prism:doi"
[37] "ot:studyId" "prism:pageRange"
[39] "dc:publisher" "ot:studyPublication"
[41] "prism:volume" "tb:title.study"
[43] "ot:agents" "generator"
[45] "prism:publicationDate" "ot:tag"
[47] "ot:comment" "ot:focalClade"
[49] "prism:startingPage" "xhtml:license"
[51] "prism:creationDate" "version"
[53] "dcterms:bibliographicCitation"
$tree_properties
[1] "ot:messages" "xsi:type"
[3] "ot:nearestTaxonMRCAName" "meta"
[5] "ot:specifiedRoot" "ot:reasonsToExcludeFromSynthesis"
[7] "tb:quality.tree" "ot:branchLengthTimeUnit"
[9] "ot:nodeLabelMode" "ot:rootNodeId"
[11] "ot:inGroupClade" "ot:ottTaxonName"
[13] "ot:branchLengthDescription" "ot:studyId"
[15] "ot:MRCAName" "ot:unrootedTree"
[17] "tb:kind.tree" "tb:type.tree"
[19] "edgeBySourceId" "ot:nodeLabelDescription"
[21] "nodeById" "ot:curatedType"
[23] "ot:nearestTaxonMRCAOttId" "ot:tag"
[25] "rootedge" "label"
[27] "ntips" "tb:ntax.tree"
[29] "ot:ottId" "ot:nodeLabelTimeUnit"
[31] "ot:outGroupEdge" "ot:branchLengthMode"
[33] "ot:MRCAOttId"
As you can see, the actual values that this properties can take are not available in the output of the function. Go to the phylesystem API wiki to get them, along with an explanation of their meaning.
To get all trees with branch lengths poprotional to time we need the function
studies_find_trees(), using the property “ot:branchLengthMode” and the value “ot:time”.
It takes some time for it to get all the information, so we will not do it now.
Go to Instructor Notes later for more information on how to do this.
Search the opentree chronogram database from datelife
In the package datelife, we have implemented a workflow that extracts all studies containing information from at least two taxa.
You can get all source chronograms from an induced subtree, as long as the tip labels are in the “name” format (and not the default “name_and_id”).
datelife takes as input either a tree with tip labels as scientific names (andd not names and ids), or a vector of scientific names.
Get a Canis subtree with tip labels that do not contain the OTT id.
canis_node_subtree <- rotl::tol_subtree(node_id = canis_node_info$node_id, label = "name")
canis_node_subtree
Phylogenetic tree with 85 tips and 28 internal nodes.
Tip labels:
Canis_lupus_pallipes, Canis_lupus_chanco, Canis_lupus_baileyi, Canis_lupus_laniger, Canis_lupus_hattai, Canis_lupus_desertorum, ...
Node labels:
, , , , , , ...
Unrooted; no branch lengths.
Now, you can use that tree as input for the get_datelife_result() function.
canis_dr <- datelife::get_datelife_result(canis_node_subtree)
We have now a list of matrices storing time of lineage divergence data for all taxon pairs.
Lists are named by the study citation, so we have that information handy at all times.
Let’s explore the output.
names(canis_dr)
[1] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[2] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[3] "Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512"
[4] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[5] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[6] "Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12"
[7] "Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845"
canis_dr[1] # look at the first element of the list
$`Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512`
Canis rufus Canis latrans Canis simensis Canis adustus
Canis rufus 0.0 2.8 2.8 3.2
Canis latrans 2.8 0.0 2.8 3.2
Canis simensis 2.8 2.8 0.0 3.2
Canis adustus 3.2 3.2 3.2 0.0
Canis aureus 3.2 3.2 3.2 2.6
Lycalopex culpaeus 6.4 6.4 6.4 6.4
Lycalopex griseus 6.4 6.4 6.4 6.4
Lycalopex gymnocercus 6.4 6.4 6.4 6.4
Lycalopex sechurae 6.4 6.4 6.4 6.4
Lycalopex vetulus 6.4 6.4 6.4 6.4
Atelocynus microtis 6.4 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4 6.4
Chrysocyon brachyurus 6.4 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8 14.8
Canis aureus Lycalopex culpaeus Lycalopex griseus
Canis rufus 3.2 6.4 6.4
Canis latrans 3.2 6.4 6.4
Canis simensis 3.2 6.4 6.4
Canis adustus 2.6 6.4 6.4
Canis aureus 0.0 6.4 6.4
Lycalopex culpaeus 6.4 0.0 1.0
Lycalopex griseus 6.4 1.0 0.0
Lycalopex gymnocercus 6.4 1.0 1.0
Lycalopex sechurae 6.4 1.0 1.0
Lycalopex vetulus 6.4 1.4 1.4
Atelocynus microtis 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4
Chrysocyon brachyurus 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8
Lycalopex gymnocercus Lycalopex sechurae
Canis rufus 6.4 6.4
Canis latrans 6.4 6.4
Canis simensis 6.4 6.4
Canis adustus 6.4 6.4
Canis aureus 6.4 6.4
Lycalopex culpaeus 1.0 1.0
Lycalopex griseus 1.0 1.0
Lycalopex gymnocercus 0.0 1.0
Lycalopex sechurae 1.0 0.0
Lycalopex vetulus 1.4 1.4
Atelocynus microtis 6.4 6.4
Cerdocyon thous 6.4 6.4
Chrysocyon brachyurus 6.4 6.4
Lycaon pictus 6.4 6.4
Speothos venaticus 6.4 6.4
Vulpes ferrilata 14.8 14.8
Lycalopex vetulus Atelocynus microtis Cerdocyon thous
Canis rufus 6.4 6.4 6.4
Canis latrans 6.4 6.4 6.4
Canis simensis 6.4 6.4 6.4
Canis adustus 6.4 6.4 6.4
Canis aureus 6.4 6.4 6.4
Lycalopex culpaeus 1.4 6.4 6.4
Lycalopex griseus 1.4 6.4 6.4
Lycalopex gymnocercus 1.4 6.4 6.4
Lycalopex sechurae 1.4 6.4 6.4
Lycalopex vetulus 0.0 6.4 6.4
Atelocynus microtis 6.4 0.0 6.4
Cerdocyon thous 6.4 6.4 0.0
Chrysocyon brachyurus 6.4 6.4 6.4
Lycaon pictus 6.4 6.4 6.4
Speothos venaticus 6.4 6.4 6.4
Vulpes ferrilata 14.8 14.8 14.8
Chrysocyon brachyurus Lycaon pictus Speothos venaticus
Canis rufus 6.4 6.4 6.4
Canis latrans 6.4 6.4 6.4
Canis simensis 6.4 6.4 6.4
Canis adustus 6.4 6.4 6.4
Canis aureus 6.4 6.4 6.4
Lycalopex culpaeus 6.4 6.4 6.4
Lycalopex griseus 6.4 6.4 6.4
Lycalopex gymnocercus 6.4 6.4 6.4
Lycalopex sechurae 6.4 6.4 6.4
Lycalopex vetulus 6.4 6.4 6.4
Atelocynus microtis 6.4 6.4 6.4
Cerdocyon thous 6.4 6.4 6.4
Chrysocyon brachyurus 0.0 6.4 6.4
Lycaon pictus 6.4 0.0 6.4
Speothos venaticus 6.4 6.4 0.0
Vulpes ferrilata 14.8 14.8 14.8
Vulpes ferrilata
Canis rufus 14.8
Canis latrans 14.8
Canis simensis 14.8
Canis adustus 14.8
Canis aureus 14.8
Lycalopex culpaeus 14.8
Lycalopex griseus 14.8
Lycalopex gymnocercus 14.8
Lycalopex sechurae 14.8
Lycalopex vetulus 14.8
Atelocynus microtis 14.8
Cerdocyon thous 14.8
Chrysocyon brachyurus 14.8
Lycaon pictus 14.8
Speothos venaticus 14.8
Vulpes ferrilata 0.0
canis_dr[length(canis_dr)] # look at the last element of the list
$`Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845`
Chrysocyon brachyurus Lycaon pictus Speothos venaticus
Chrysocyon brachyurus 0.00000 13.58235 13.58235
Lycaon pictus 13.58235 0.00000 10.91794
Speothos venaticus 13.58235 10.91794 0.00000
Lycalopex vetulus 16.55066 16.55066 16.55066
Lycalopex fulvipes 16.55066 16.55066 16.55066
Lycalopex culpaeus 16.55066 16.55066 16.55066
Lycalopex gymnocercus 16.55066 16.55066 16.55066
Lycalopex griseus 16.55066 16.55066 16.55066
Lycalopex sechurae 16.55066 16.55066 16.55066
Cerdocyon thous 16.55066 16.55066 16.55066
Atelocynus microtis 16.55066 16.55066 16.55066
Canis adustus 16.55066 16.55066 16.55066
Canis latrans 16.55066 16.55066 16.55066
Canis aureus 16.55066 16.55066 16.55066
Canis simensis 16.55066 16.55066 16.55066
Dusicyon australis 18.25725 18.25725 18.25725
Vulpes ferrilata 25.20000 25.20000 25.20000
Lycalopex vetulus Lycalopex fulvipes Lycalopex culpaeus
Chrysocyon brachyurus 16.550658 16.550658 16.550657
Lycaon pictus 16.550658 16.550658 16.550657
Speothos venaticus 16.550658 16.550658 16.550657
Lycalopex vetulus 0.000000 3.064400 5.618639
Lycalopex fulvipes 3.064400 0.000000 5.618639
Lycalopex culpaeus 5.618639 5.618639 0.000000
Lycalopex gymnocercus 5.618639 5.618639 2.856292
Lycalopex griseus 5.618640 5.618640 3.180365
Lycalopex sechurae 5.618640 5.618640 3.414349
Cerdocyon thous 8.074640 8.074640 8.074639
Atelocynus microtis 8.604404 8.604404 8.604403
Canis adustus 12.761959 12.761959 12.761958
Canis latrans 12.761958 12.761958 12.761957
Canis aureus 12.761958 12.761958 12.761957
Canis simensis 12.761958 12.761958 12.761957
Dusicyon australis 18.257250 18.257250 18.257249
Vulpes ferrilata 25.200000 25.200000 25.199999
Lycalopex gymnocercus Lycalopex griseus
Chrysocyon brachyurus 16.550657 16.550658
Lycaon pictus 16.550657 16.550658
Speothos venaticus 16.550657 16.550658
Lycalopex vetulus 5.618639 5.618640
Lycalopex fulvipes 5.618639 5.618640
Lycalopex culpaeus 2.856292 3.180365
Lycalopex gymnocercus 0.000000 3.180365
Lycalopex griseus 3.180365 0.000000
Lycalopex sechurae 3.414349 3.414350
Cerdocyon thous 8.074639 8.074640
Atelocynus microtis 8.604403 8.604404
Canis adustus 12.761958 12.761959
Canis latrans 12.761957 12.761958
Canis aureus 12.761957 12.761958
Canis simensis 12.761957 12.761958
Dusicyon australis 18.257249 18.257250
Vulpes ferrilata 25.199999 25.200000
Lycalopex sechurae Cerdocyon thous Atelocynus microtis
Chrysocyon brachyurus 16.550658 16.550658 16.550658
Lycaon pictus 16.550658 16.550658 16.550658
Speothos venaticus 16.550658 16.550658 16.550658
Lycalopex vetulus 5.618640 8.074640 8.604404
Lycalopex fulvipes 5.618640 8.074640 8.604404
Lycalopex culpaeus 3.414349 8.074639 8.604403
Lycalopex gymnocercus 3.414349 8.074639 8.604403
Lycalopex griseus 3.414350 8.074640 8.604404
Lycalopex sechurae 0.000000 8.074640 8.604404
Cerdocyon thous 8.074640 0.000000 8.604404
Atelocynus microtis 8.604404 8.604404 0.000000
Canis adustus 12.761959 12.761959 12.761959
Canis latrans 12.761958 12.761958 12.761958
Canis aureus 12.761958 12.761958 12.761958
Canis simensis 12.761958 12.761958 12.761958
Dusicyon australis 18.257250 18.257250 18.257250
Vulpes ferrilata 25.200000 25.200000 25.200000
Canis adustus Canis latrans Canis aureus Canis simensis
Chrysocyon brachyurus 16.55066 16.55066 16.55066 16.55066
Lycaon pictus 16.55066 16.55066 16.55066 16.55066
Speothos venaticus 16.55066 16.55066 16.55066 16.55066
Lycalopex vetulus 12.76196 12.76196 12.76196 12.76196
Lycalopex fulvipes 12.76196 12.76196 12.76196 12.76196
Lycalopex culpaeus 12.76196 12.76196 12.76196 12.76196
Lycalopex gymnocercus 12.76196 12.76196 12.76196 12.76196
Lycalopex griseus 12.76196 12.76196 12.76196 12.76196
Lycalopex sechurae 12.76196 12.76196 12.76196 12.76196
Cerdocyon thous 12.76196 12.76196 12.76196 12.76196
Atelocynus microtis 12.76196 12.76196 12.76196 12.76196
Canis adustus 0.00000 10.31455 10.31455 10.31455
Canis latrans 10.31455 0.00000 4.44640 6.60000
Canis aureus 10.31455 4.44640 0.00000 6.60000
Canis simensis 10.31455 6.60000 6.60000 0.00000
Dusicyon australis 18.25725 18.25725 18.25725 18.25725
Vulpes ferrilata 25.20000 25.20000 25.20000 25.20000
Dusicyon australis Vulpes ferrilata
Chrysocyon brachyurus 18.25725 25.2
Lycaon pictus 18.25725 25.2
Speothos venaticus 18.25725 25.2
Lycalopex vetulus 18.25725 25.2
Lycalopex fulvipes 18.25725 25.2
Lycalopex culpaeus 18.25725 25.2
Lycalopex gymnocercus 18.25725 25.2
Lycalopex griseus 18.25725 25.2
Lycalopex sechurae 18.25725 25.2
Cerdocyon thous 18.25725 25.2
Atelocynus microtis 18.25725 25.2
Canis adustus 18.25725 25.2
Canis latrans 18.25725 25.2
Canis aureus 18.25725 25.2
Canis simensis 18.25725 25.2
Dusicyon australis 0.00000 25.2
Vulpes ferrilata 25.20000 0.0
Get your chronograms
Then, it is really easy to go from a matrix to a tree, using the function summarize_datelife_result() with the option summary_format = "phylo_all". Note the printed output returns a summary of taxa that have branch length information in the database.
canis_phylo_all <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_all")
Source chronograms from:
1: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
2: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
3: Bininda-Emonds, Olaf R. P., Marcel Cardillo, Kate E. Jones, Ross D. E. MacPhee, Robin M. D. Beck, Richard Grenyer, Samantha A. Price, Rutger A. Vos, John L. Gittleman, Andy Purvis. 2007. The delayed rise of present-day mammals. Nature 446 (7135): 507-512
4: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
5: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
6: Nyakatura, Katrin, Olaf RP Bininda-Emonds. 2012. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biology 10 (1): 12
7: Hedges, S. Blair, Julie Marin, Michael Suleski, Madeline Paymer, Sudhir Kumar. 2015. Tree of life reveals clock-like speciation and diversification. Molecular Biology and Evolution 32 (4): 835-845
Input taxa presence across source chronograms:
taxon chronograms
1 Canis rufus 3/7
2 Canis latrans 7/7
3 Canis simensis 7/7
4 Canis adustus 7/7
5 Canis aureus 7/7
6 Lycalopex culpaeus 7/7
7 Lycalopex griseus 7/7
8 Lycalopex gymnocercus 7/7
9 Lycalopex sechurae 7/7
10 Lycalopex vetulus 7/7
11 Atelocynus microtis 7/7
12 Cerdocyon thous 7/7
13 Chrysocyon brachyurus 7/7
14 Lycaon pictus 7/7
15 Speothos venaticus 7/7
16 Vulpes ferrilata 7/7
17 Dusicyon australis 4/7
18 Lycalopex fulvipes 4/7
Input taxa completely absent from source chronograms:
taxon
1 Canis himalayensis
2 Canis indica
3 Canis environmental sample
4 Canis anthus
5 Canis antarticus
6 Canis dingo
7 Canis ameghinoi
8 Canis argentinus
9 Canis cautleyi
10 Canis chanco
11 Canis chrysurus
12 Canis curvipalatus
13 Canis dukhunensis
14 Canis etruscus
15 Canis gezi
16 Canis himalaicus
17 Canis kokree
18 Canis lanka
19 Canis lateralis
20 Canis naria
21 Canis nehringi
22 Canis pallipes
23 Canis palustris
24 Canis peruanus
25 Canis primaevus
26 Canis sladeni
27 Canis tarijensis
28 Canis ursinus
29 Canis ferox
30 Canis lupus pallipes
31 Canis lupus chanco
32 Canis lupus baileyi
33 Canis lupus laniger
34 Canis lupus hattai
35 Canis lupus desertorum
36 Canis lupus familiaris
37 Canis lupus mogollonensis
38 Canis lupus hodophilax
39 Canis lupus dingo
40 Canis lupus labradorius
41 Canis lupus signatus
42 Canis lupus lupus
43 Canis lupus lycaon
44 Canis lupus lupaster
45 Canis lupus campestris
46 Canis lupus arctos
47 Canis lupus variabilis
48 Canis lupus orion
49 Canis dirus
50 Canis armbrusteri
51 Speothos pacivorus
52 Cuon primaevus
53 Cuon javanicus
54 Cuon stehlini
55 Cuon alpinus lepturus
56 Canis edwardii
57 Canis lepophagus
58 Canis cedazoensis
59 Canis davisi
60 Dusicyon avus
61 Dusicyon darwini
62 Dusicyon gymnocercus
63 Dusicyon proplatensis
64 Lycalopex sp. Fuegian dog
65 Lycalopex fulvicaudus
66 Canis mesomelas elongae
67 Cerdocyon ensenadensis
Plot your results
To plot the resulting tree, you can use the plot.phylo() function from ape.
You can also use the function plot_phylo_all(), that adds the study citation as title and a geostratigraphic axis.
datelife::plot_phylo_all(trees = canis_phylo_all)






Key Points
datelife stores all chronograms from the Open Tree of Life phylesystem.
chronograms are stored in the
opentree_chronogramsobject.source chronograms are retrieved at the species level only (for now).
7. Summarizing branch length information
Overview
Teaching: 5 min
Exercises: 5 minQuestions
How do I summarize information from different source chronograms?
How do I choose a preferred source chronogram?
Objectives
Understanding the depth of uncertainty around age estimates.
Now that we have a collection of chronograms containing our taxa of interest, we can go on to summarize the information in them.
There is no consensus on the best way to do this.
We have implemented two ways of summarizing information from several chronograms into a single one. The fastest one is using the median of node ages for each node with available information, and then evenly distributing ages across nodes.
canis_phylo_median <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_median")
Check that we actually went from a list of matrices to a tree with branch lengths:
canis_phylo_median
Phylogenetic tree with 18 tips and 13 internal nodes.
Tip labels:
Canis_rufus, Canis_simensis, Speothos_venaticus, Lycaon_pictus, Canis_latrans, Canis_aureus, ...
Node labels:
n1, n2, n3, n4, n5, n6, ...
Unrooted; includes branch lengths.
Good. Now we can plot our chronogram!
ape::plot.phylo(canis_phylo_median, cex = 1.2)
# Add the time axis:
ape::axisPhylo()
# And a little hack to add the axis name:
graphics::mtext("Time (myrs)", side = 1, line = 2, at = max(get("last_plot.phylo",envir = .PlotPhyloEnv)$xx) * 0.5)
Challenge! Get the other type of summary chronogram
Hint: Explore options from the argument summary_format in the function
summarize_datelife_result()Solution
canis_phylo_sdm <- datelife::summarize_datelife_result(canis_dr, summary_format = "phylo_sdm")canis_phylo_sdmPhylogenetic tree with 18 tips and 13 internal nodes. Tip labels: Canis_rufus, Canis_simensis, Speothos_venaticus, Lycaon_pictus, Canis_latrans, Canis_aureus, ... Node labels: n1, n2, n3, n4, n5, n6, ... Unrooted; includes branch lengths.ape::plot.phylo(canis_phylo_sdm, cex = 1.2) ape::axisPhylo() graphics::mtext("Time (myrs)", side = 1, line = 2, at = max(get("last_plot.phylo",envir = .PlotPhyloEnv)$xx) * 0.5)
As you can note, the SDM sumary chronogram is slightly older than the median summary chronogram!
Finally, give it a try on the web browser of datelife, too. You can do the same things using a graphical user interface. It is fun!
Key Points
Source chronograms have a wide range of variation in age estimates.